4JCA
 
 | | Crystal Structure of the apo form of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1. Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | CITRIC ACID, RUBIDIUM ION, serine hydrolase | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JLL
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1 covalently bound with FP-alkyne, Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
1QYS
 
 | | Crystal structure of Top7: A computationally designed protein with a novel fold | | Descriptor: | TOP7 | | Authors: | Kuhlman, B, Dantas, G, Ireton, G.C, Varani, G, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-09-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
Science, 302, 2003
|
|
2L9V
 
 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
2MJZ
 
 | |
2MDW
 
 | |
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2LPZ
 
 | | Atomic model of the Type-III Secretion System Needle | | Descriptor: | Protein prgI | | Authors: | Loquet, A, Sgourakis, N.G, Gupta, R, Giller, K, Riedel, D, Goosmann, C, Griesinger, C, Kolbe, M.G, Baker, D, Becker, S, Lange, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of the type III secretion system needle.
Nature, 486, 2012
|
|
2MBM
 
 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
1PVH
 
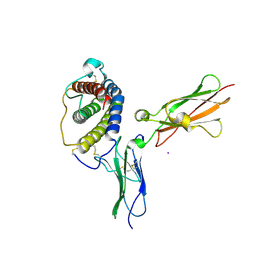 | | Crystal structure of leukemia inhibitory factor in complex with gp130 | | Descriptor: | IODIDE ION, Interleukin-6 receptor beta chain, Leukemia inhibitory factor | | Authors: | Boulanger, M.J, Bankovich, A.J, Kortemme, T, Baker, D, Garcia, K.C. | | Deposit date: | 2003-06-27 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130.
Mol.Cell, 12, 2003
|
|
2MDV
 
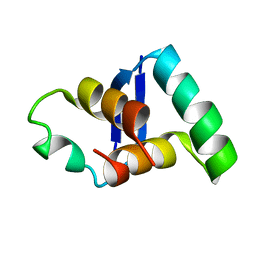 | |
2MV0
 
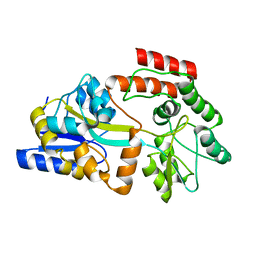 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | Descriptor: | Rossmann 2x3 fold protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
2MRA
 
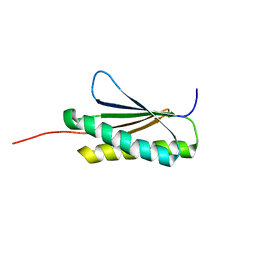 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | Descriptor: | De novo designed protein OR459 | | Authors: | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
2KS6
 
 | |
4IX0
 
 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | NICKEL (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZ6
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JML
 
 | | Conversion of Monomeric Protein L to an Obligate Dimer by Computational Protein Design | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y.J, Baker, D. | | Deposit date: | 2001-07-19 | | Release date: | 2001-10-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conversion of monomeric protein L to an obligate dimer by computational protein design.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4IWW
 
 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | COBALT (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
1K50
 
 | | A V49A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K53
 
 | | Monomeric Protein L B1 Domain with a G15A Mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K52
 
 | | Monomeric Protein L B1 Domain with a K54G mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
4J9T
 
 | | Crystal structure of a putative, de novo designed unnatural amino acid dependent metalloprotein, northeast structural genomics consortium target OR61 | | Descriptor: | ARSENIC, GLYCEROL, designed unnatural amino acid dependent metalloprotein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Mills, J.H, Khare, S.D, Everett, J.K, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
