7FV5
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z606695272 | | Descriptor: | 4-(furan-2-carbonyl)-N-(pyridin-2-yl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV0
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z44602337 | | Descriptor: | 4-(furan-2-carbonyl)-N-phenylpiperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV6
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1334218055 | | Descriptor: | N-methyl-4-[5-(phenoxymethyl)furan-2-carbonyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVH
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z5067911819 | | Descriptor: | (2R)-4-(furan-3-carbonyl)-N-(4-methoxyphenyl)-2-methylpiperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FUS
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z44602363 | | Descriptor: | N-(2-chlorophenyl)-4-(furan-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FUU
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z445977856 | | Descriptor: | N-{[(2S)-oxolan-2-yl]methyl}-4-(thiophene-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVK
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z409964562 | | Descriptor: | 4-(furan-2-carbonyl)-N-(2-methoxy-5-methylphenyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV7
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1929967066 | | Descriptor: | (3R)-4-(furan-2-carbonyl)-3-methyl-N-(propan-2-yl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FV8
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z964297186 | | Descriptor: | 4-(3-chlorobenzoyl)-N-[3-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-3-oxopropyl]-1,4-diazepane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVD
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z1424453050 | | Descriptor: | 4-(thieno[3,2-b]thiophene-2-carbonyl)-N-[(2S)-2,3,3-trimethylbutyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7FVE
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z488932160 | | Descriptor: | 4-(5-bromofuran-2-carbonyl)-N-[3-(3-methylphenoxy)propyl]piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1A7H
 
 | | GAMMA S CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMAS CRYSTALLIN | | Authors: | Basak, A.K, Slingsby, C. | | Deposit date: | 1998-03-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The C-terminal domains of gammaS-crystallin pair about a distorted twofold axis.
Protein Eng., 11, 1998
|
|
1BD7
 
 | | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Descriptor: | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Authors: | Wright, G, Basak, A.K, Mayr, E.-M, Glockshuber, R, Slingsby, C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circular permutation of betaB2-crystallin changes the hierarchy of domain assembly.
Protein Sci., 7, 1998
|
|
1A5D
 
 | | GAMMAE CRYSTALLIN FROM RAT LENS | | Descriptor: | GAMMAE CRYSTALLIN | | Authors: | Norledge, B.V, Hay, R, Bateman, O.A, Slingsby, C, Driessen, H.P.C. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Towards a molecular understanding of phase separation in the lens: a comparison of the X-ray structures of two high Tc gamma-crystallins, gammaE and gammaF, with two low Tc gamma-crystallins, gammaB and gammaD.
Exp.Eye Res., 65, 1997
|
|
2J9I
 
 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | Descriptor: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | Deposit date: | 2006-11-09 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
2KG2
 
 | | Solution structure of a PDZ protein | | Descriptor: | Tax1-binding protein 3 | | Authors: | Durney, M.A, Birrane, G, Anklin, C, Soni, A, Ladias, J.A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human Tax-interacting protein-1.
J.Biomol.Nmr, 45, 2009
|
|
1DSL
 
 | | GAMMA B CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
1E7N
 
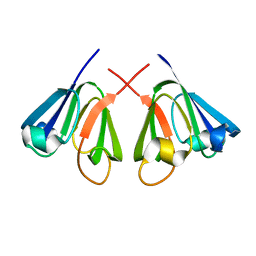 | | The N-terminal domain of beta-B2-crystallin resembles the putative ancestral homodimer | | Descriptor: | BETA-CRYSTALLIN B2 | | Authors: | Clout, N.J, Basak, A, Wieligmann, K, Bateman, O.A, Jaenicke, R, Slingsby, C. | | Deposit date: | 2000-08-31 | | Release date: | 2000-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The N-Terminal Domain of Betab2-Crystallin Resembles the Putative Ancestral Homodimer.
J.Mol.Biol., 304, 2000
|
|
2MM8
 
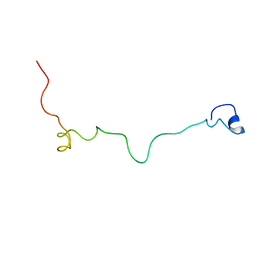 | | Structural and biochemical characterization of Jaburetox | | Descriptor: | Urease | | Authors: | Dobrovolska, O, Lopez, F, Ciurli, S, Zambelli, B, Carlini, C. | | Deposit date: | 2014-03-12 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, 15N resonance assignments of reduced Jaburetox
To be Published
|
|
2O20
 
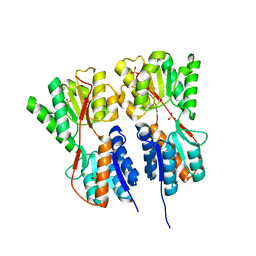 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | Descriptor: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | Authors: | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4IGU
 
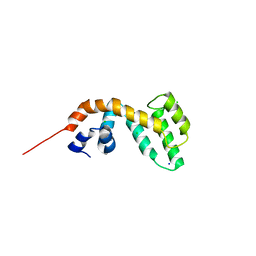 | |
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG8
 
 | |
8IG7
 
 | |
8IG9
 
 | |
