4NG7
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Citrobacter koseri (CKO_04899), Target EFI-510094, apo, open structure | | Descriptor: | TRAP periplasmic solute binding protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NT0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate | | Descriptor: | 4-hydroxy-1-(5-O-phosphono-beta-D-ribofuranosyl)pyridin-2(1H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 3-deazauridine 5'-monophosphate
To be Published
|
|
4NN3
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens (Desal_2161), Target EFI-510109, with bound orotic acid | | Descriptor: | CHLORIDE ION, OROTIC ACID, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-16 | | Release date: | 2013-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NQ8
 
 | | Crystal structure of a trap periplasmic solute binding protein from Bordetella bronchispeptica (bb3421), target EFI-510039, with density modeled as pantoate | | Descriptor: | CHLORIDE ION, PANTOATE, Putative periplasmic substrate-binding transport protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-24 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NSN
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry | | Descriptor: | ADENINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 030972, orthorhombic symmetry.
To be Published
|
|
4NX5
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-AZA URIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate
To be Published
|
|
4NS1
 
 | | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Porphyromonas gingivalis ATCC 33277, NYSGRC Target 30972.
To be Published
|
|
4NX1
 
 | | Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-talopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O11
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-14 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-hydroxyuridine 5'-monophosphate
To be Published
|
|
4MAR
 
 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate. | | Descriptor: | MAGNESIUM ION, Purine nucleoside phosphorylase DeoD-type, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279 complexed with sulfate.
To be Published
|
|
4M3N
 
 | | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804. | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase from Meiothermus ruber DSM 1279, NYSGRC Target 029804.
To be Published
|
|
4M6D
 
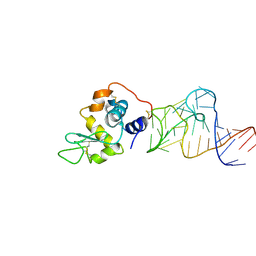 | | Crystal structure of the aptamer minF-lysozyme complex. | | Descriptor: | Lysozyme C, aptamer | | Authors: | Malashkevich, V.N, Padlan, F.C, Toro, R, Girvin, M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of the aptamer minF-lysozyme complex.
To be Published
|
|
4LZA
 
 | | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. | | Descriptor: | Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700.
To be Published
|
|
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F62
 
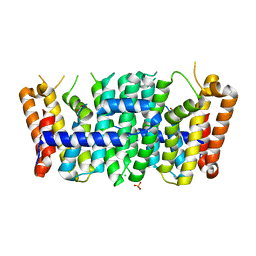 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4O94
 
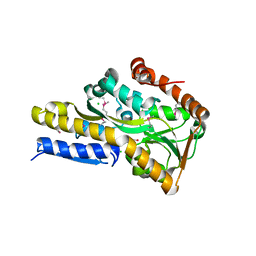 | | Crystal structure of a trap periplasmic solute binding protein from Rhodopseudomonas palustris HaA2 (RPB_3329), Target EFI-510223, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4M1E
 
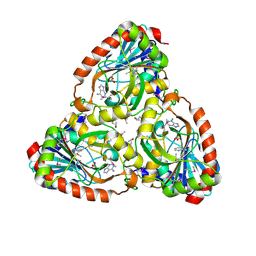 | | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364. | | Descriptor: | ADENINE, PYRIDINE-2-CARBOXYLIC ACID, Purine nucleoside phosphorylase, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of purine nucleoside phosphorylase I from Planctomyces limnophilus DSM 3776, NYSGRC Target 029364.
To be Published
|
|
4MHF
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Glucuronate, space group P21 | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit, alpha-D-glucopyranuronic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NUW
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate
To be Published
|
|
4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
4OVQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS, TARGET EFI-510230, WITH BOUND BETA-D-GLUCURONATE | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate ABC transporter, substrate-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-25 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4OVP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1, TARGET EFI-510292, WITH BOUND ALPHA-D-MANURONATE | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-mannopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O8M
 
 | | Crystal structure of a trap periplasmic solute binding protein actinobacillus succinogenes 130z, target EFI-510004, with bound L-galactonate | | Descriptor: | CHLORIDE ION, L-galactonic acid, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-28 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4NHW
 
 | | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275, with one glutathione bound per one protein subunit | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275
To be Published
|
|
