6JG2
 
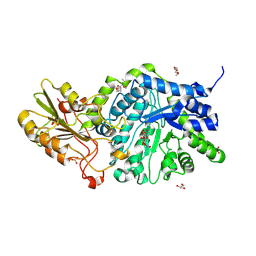 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGD
 
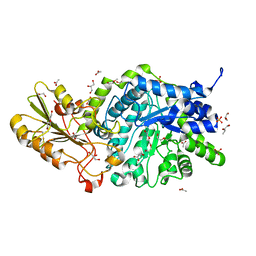 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGP
 
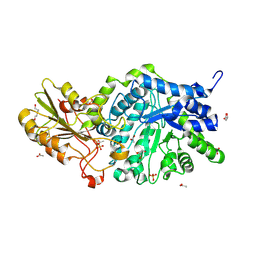 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2W3M
 
 | |
6JGA
 
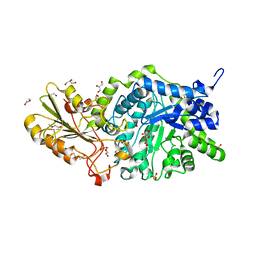 | | Crystal structure of barley exohydrolaseI W286F in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-oxane-2,3,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JGL
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with methyl 2-thio-beta-sophoroside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2V5V
 
 | | W57E Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, MAGNESIUM ION | | Authors: | Herguedas, B, Martinez-Julvez, M, Perez-Dorado, I, Goni, G, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
6JVN
 
 | | Crystal structure of human MTH1 in complex with compound MI1020 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-methyl-6-morpholin-4-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
2W3B
 
 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR MYCOBACTERIUM AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | | Descriptor: | 6-{[(2,5-DIETHOXYPHENYL)AMINO]METHYL}-5-METHYLPYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, FOLIC ACID, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Borhani, D.W. | | Deposit date: | 2008-11-11 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium Avium Dihydrofolate Reductase by a Lipophilic Antifolate
To be Published
|
|
2VQD
 
 | |
6JVS
 
 | | Crystal structure of human MTH1 in complex with compound MI1029 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-[4-(oxetan-3-yl)piperazin-1-yl]pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
2W3W
 
 | | MYCOBACTERIUM AVIUM DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND A LIPOPHILIC ANTIFOLATE SELECTIVE FOR M. AVIUM DHFR, 6-((2,5- DIETHOXYPHENYL)AMINOMETHYL)-2,4-DIAMINO-5-METHYLPYRIDO(2,3-D) PYRIMIDINE (SRI-8686) | | Descriptor: | 6-{[(2,5-DIETHOXYPHENYL)AMINO]METHYL}-5-METHYLPYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Leung, A.K.W, Reynolds, R.C, Borhani, D.W. | | Deposit date: | 2008-11-16 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Selective Inhibition of Mycobacterium Avium Dihydrofolate Reductase by a Lipophilic Antifolate
To be Published
|
|
6IGZ
 
 | | Structure of PSI-LHCI | | Descriptor: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Xiong, P, Xiaochun, Q. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
6HP5
 
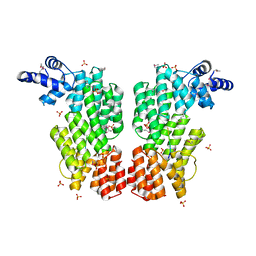 | | ARBITRIUM PEPTIDE RECEPTOR FROM SPBETA PHAGE | | Descriptor: | GLY-MET-PRO-ARG-GLY-ALA, SPBc2 prophage-derived uncharacterized protein YopK, SULFATE ION | | Authors: | Marina, A, Gallego del Sol, F. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Deciphering the Molecular Mechanism Underpinning Phage Arbitrium Communication Systems.
Mol.Cell, 74, 2019
|
|
6HML
 
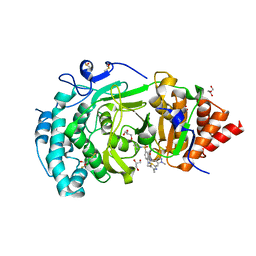 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00017299 | | Descriptor: | 1-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-3-[(1-methylpyrazol-4-yl)methyl]quinazoline-2,4-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Barkauskaite, E. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HP3
 
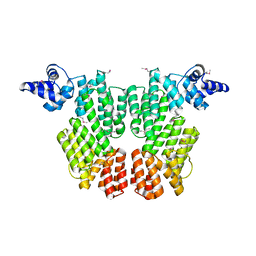 | |
2W08
 
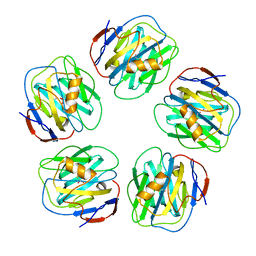 | | The structure of serum amyloid P component bound to 0-phospho- threonine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHOTHREONINE, ... | | Authors: | Kolstoe, S.E, Pepys, M.B, Wood, S.P. | | Deposit date: | 2008-08-12 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Dissection of Alzheimer'S Disease Neuropathology by Depletion of Serum Amyloid P Component.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1BL6
 
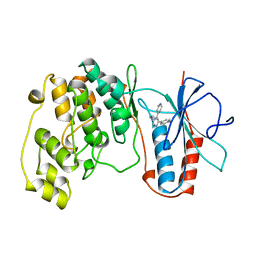 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
6HMM
 
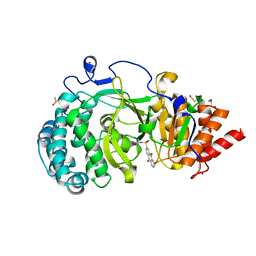 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00013907 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HP7
 
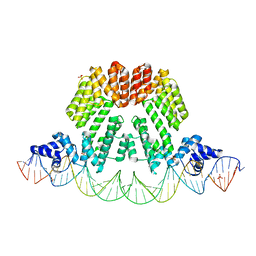 | |
6I3U
 
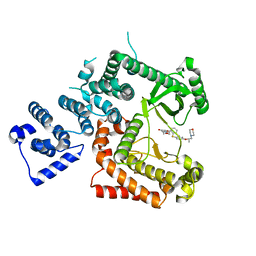 | |
2W3V
 
 | |
2VPQ
 
 | |
