8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8GDW
 
 | |
8GBK
 
 | |
8GF4
 
 | |
8HO1
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant F387G | | Descriptor: | CALCIUM ION, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNZ
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 6FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(5-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNY
 
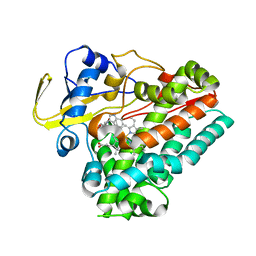 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 5FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(4-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7DZ9
 
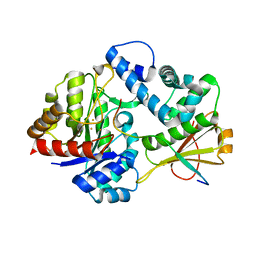 | | MbnABC complex | | Descriptor: | FE (III) ION, MbnA, MbnB, ... | | Authors: | Chao, D, Dan, Z, Yijun, G, Wei, C. | | Deposit date: | 2021-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7V3Z
 
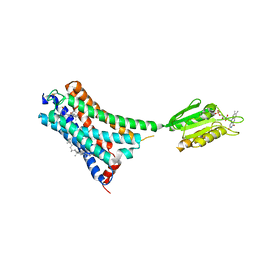 | | Structure of cannabinoid receptor type 1(CB1) | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, CHOLESTEROL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liu, Z.J, Shen, L, Hua, T, Yao, D.Q, Wu, L.J. | | Deposit date: | 2021-08-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A Genetically Encoded F-19 NMR Probe Reveals the Allosteric Modulation Mechanism of Cannabinoid Receptor 1.
J.Am.Chem.Soc., 143, 2021
|
|
7XZH
 
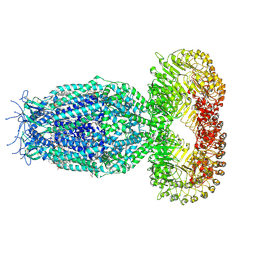 | | Cryo-EM structure of human LRRC8A | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Liu, H, Liao, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural insights into anion selectivity and activation mechanism of LRRC8 volume-regulated anion channels.
Cell Rep, 42, 2023
|
|
5YQ7
 
 | | Cryo-EM structure of the RC-LH core complex from Roseiflexus castenholzii | | Descriptor: | 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, BACTERIOCHLOROPHYLL A, ... | | Authors: | Shi, Y, Xin, Y.Y, Niu, T.X, Wang, Q.Q, Niu, W.Q, Huang, X.J, Ding, W, Blankenship, R.E, Xu, X.L, Sun, F. | | Deposit date: | 2017-11-05 | | Release date: | 2018-05-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote.
Nat Commun, 9, 2018
|
|
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Chen, R.C, Gao, S.S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
7XE8
 
 | |
5ZLU
 
 | | Ribosome Structure bound to ABC-F protein. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Su, W.X, Kumar, V, Ero, R, Andrew, S.W.W, Jian, S, Yong-Gui, G. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-01 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ribosome protection by antibiotic resistance ATP-binding cassette protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BG3
 
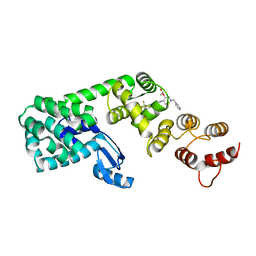 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
7EWQ
 
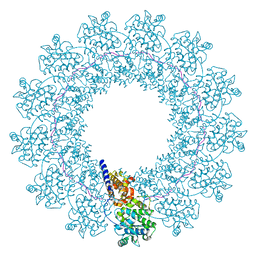 | | Structure of Mumps virus nucleocapsid ring | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Su, X, Shen, Q, Shan, H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
8J6P
 
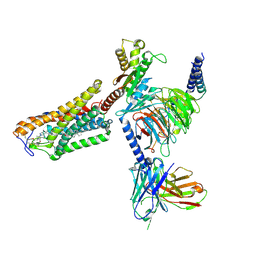 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6R
 
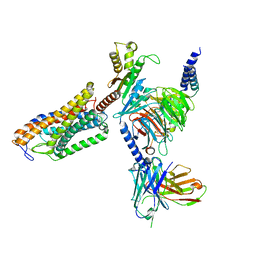 | | Cryo-EM structure of the MK-6892-bound human HCAR2-Gi1 complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8J6Q
 
 | | Cryo-EM structure of the 3-HB and compound 9n-bound human HCAR2-Gi1 complex | | Descriptor: | (3R)-3-hydroxybutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, ... | | Authors: | Mao, C, Gao, M, Zang, S, Zhu, Y, Ma, X, Zhang, Y. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Nat Commun, 14, 2023
|
|
8YFK
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFM
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
