7O4Z
 
 | | Crystal structure of the carbonic anhydrase-like domain of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carboxysome assembly protein CcmM, NICKEL (II) ION | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7O54
 
 | | Crystal structure of the carbonic anhydrase-like domain of CcmM in complex with the C-terminal 17 residues of CcaA from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, Carboxysome assembly protein CcmM, ... | | Authors: | Zang, K, Wang, H, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5GTX
 
 | | Crystal structure of mutated buckwheat glutaredoxin | | Descriptor: | buckwheat glutaredoxin | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
2WOC
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QC2
 
 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
2WOE
 
 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOD
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2Z1B
 
 | | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculs | | Descriptor: | Delta-aminolevulinic acid dehydratase | | Authors: | Xie, Y, Wang, H, Kawazoe, M, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculs
To be Published
|
|
2YY0
 
 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | Descriptor: | C-Myc-binding protein | | Authors: | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
9JAQ
 
 | | Cryo-EM structure of MPXV core protease in the apo-form | | Descriptor: | Core protease I7 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KQV
 
 | | Cryo-EM structure of MPXV core protease in complex with aloxistatin(E64d) | | Descriptor: | Core protease I7, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
9KR6
 
 | | Cryo-EM structure of MPXV core protease in complex with the substrate derivative I-G18 | | Descriptor: | Core protease I7, Core protein VP8 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 2025
|
|
3FOK
 
 | | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115 | | Descriptor: | uncharacterized protein Cgl0159 | | Authors: | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115
To be Published
|
|
6OCR
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3GAC
 
 | | Structure of mif with HPP | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, Macrophage migration inhibitory factor-like protein, ... | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3GAD
 
 | | Structure of apomif | | Descriptor: | ACETIC ACID, Macrophage migration inhibitory factor-like protein, SULFATE ION | | Authors: | Zhou, Y.-F, Su, X.-D, Shao, D, Wang, H. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional comparison of MIF ortholog from Plasmodium yoelii with MIF from its rodent host
Mol.Immunol., 47, 2010
|
|
6TMX
 
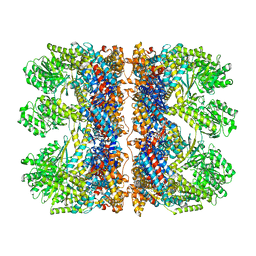 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
3E0H
 
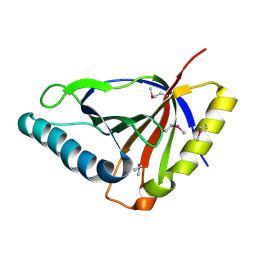 | | Crystal structure of an uncharacterized protein from Chlorobium tepidum. NorthEast Structural Genomics target CtR107. | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of an uncharacterized protein from Chlorobium tepidum. NorthEast Structural Genomics target CtR107.
To be Published
|
|
3E0E
 
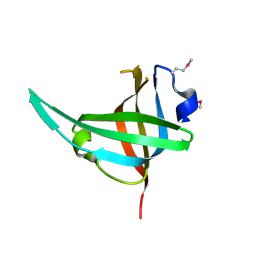 | | Crystal structure of a domain of replication protein A from Methanococcus maripaludis. NorthEast Structural Genomics targe MrR110B | | Descriptor: | Replication protein A | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a domain of replication protein A from Methanococcus maripaludis. NorthEast Structural Genomics targe MrR110B
To be Published
|
|
3E1E
 
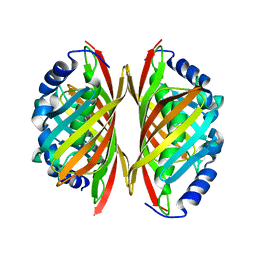 | | Crystal structure of a Thioesterase family protein from Silicibacter pomeroyi. NorthEast Structural Genomics target SiR180A | | Descriptor: | Thioesterase family protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-04 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a Thioesterase family protein from Silicibacter pomeroyi. NorthEast Structural Genomics target SiR180A
To be Published
|
|
3EN2
 
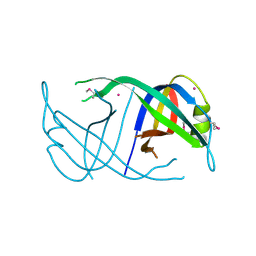 | | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C. | | Descriptor: | POTASSIUM ION, Probable primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Wang, H, Sahdev, S, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the protein priB from Ralstonia solanacearum at the resolution 2.3A. Northeast Structural Genomics Consortium target RsR213C.
To be Published
|
|
3E8P
 
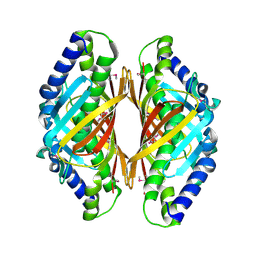 | | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246. | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Janjua, H, Ciccosanti, C, Foote, E.L, Wang, H, Xiao, R, Nair, R, Everett, J, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the protein Q8E9M7 from Shewanella oneidensis related to thioesterase superfamily. Northeast Structural Genomics Consortium target SoR246.
To be Published
|
|
3ESI
 
 | | Crystal structure of an uncharacterized protein from Erwinia carotovora subsp. atroseptica. Northeast Structural Genomics target EwR179 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Lew, S, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Erwinia carotovora subsp. atroseptica. Northeast Structural Genomics target EwR179
To be Published
|
|
