2HEB
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEA
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2HEF
 
 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
2D5V
 
 | | Crystal structure of HNF-6alpha DNA-binding domain in complex with the TTR promoter | | Descriptor: | 5'-D(*AP*TP*TP*AP*TP*TP*GP*AP*CP*TP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*AP*GP*TP*CP*AP*AP*TP*AP*AP*T)-3', ACETATE ION, ... | | Authors: | Iyaguchi, D, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2005-11-07 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA recognition mechanism of the ONECUT homeodomain of transcription factor HNF-6
Structure, 15, 2007
|
|
3GJW
 
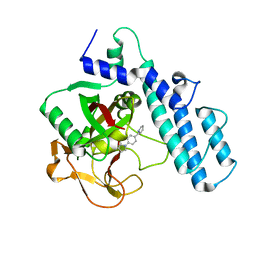 | | PARP complexed with A968427 | | Descriptor: | 7-(pyrrolidin-1-ylmethyl)pyrrolo[1,2-a]quinoxalin-4(5H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and SAR of novel tricyclic quinoxalinone inhibitors of poly(ADP-ribose)polymerase-1 (PARP-1)
Bioorg.Med.Chem.Lett., 19, 2009
|
|
9JI2
 
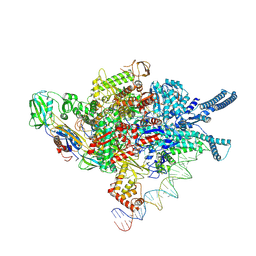 | |
9KET
 
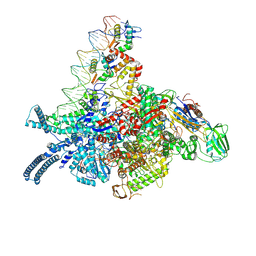 | |
9KEV
 
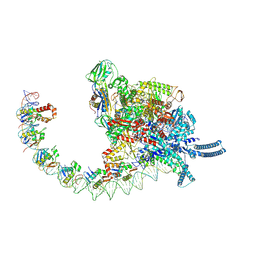 | |
9KEU
 
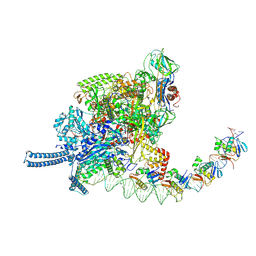 | |
3QDD
 
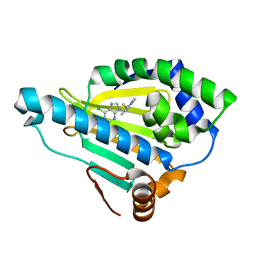 | | HSP90A N-terminal domain in complex with BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Arndt, J.W, Biamonte, M.A. | | Deposit date: | 2011-01-18 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | EC144 Is a Potent Inhibitor of the Heat Shock Protein 90.
J.Med.Chem., 55, 2012
|
|
8HML
 
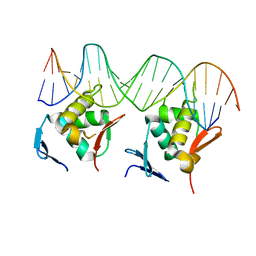 | |
2QCY
 
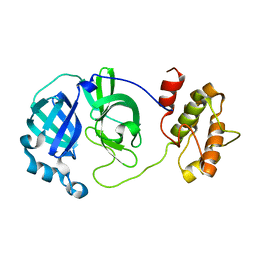 | |
5GVU
 
 | |
7VWY
 
 | |
7W5Y
 
 | |
7W5W
 
 | |
8K1Y
 
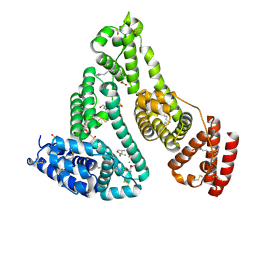 | | Crystal structure of human serum albumin and ruthenium Val complex adduct | | Descriptor: | Albumin, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gong, W.J, Xie, L.L, Wang, W.M, Wang, H.F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the anti-proliferation activity and photoinduced NO release of four nitrosylruthenium isomeric complexes and their HSA complex adducts.
Metallomics, 16, 2024
|
|
2MJV
 
 | |
7RK0
 
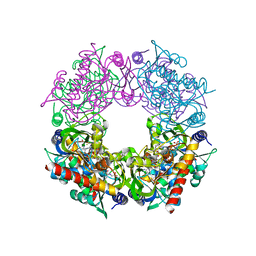 | | Crystal structure of Thermovibrio ammonificans THI4 | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (III) ION, Thiamine thiazole synthase | | Authors: | Li, Q, Bruner, S.D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and function of aerotolerant, multiple-turnover THI4 thiazole synthases.
Biochem.J., 478, 2021
|
|
6LY9
 
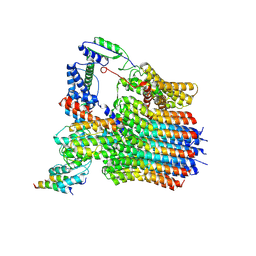 | | The membrane-embedded Vo domain of V/A-ATPase from Thermus thermophilus | | Descriptor: | V-type ATP synthase subunit C, V-type ATP synthase subunit E, V-type ATP synthase subunit I, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6LY8
 
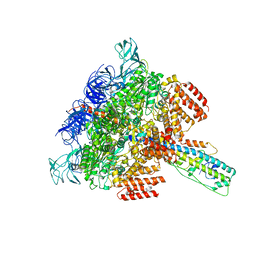 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
4XB2
 
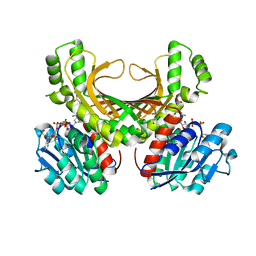 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
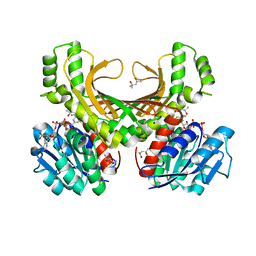 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
7XK4
 
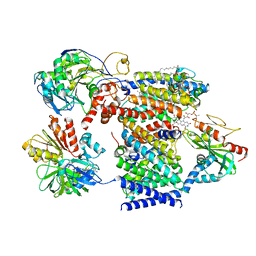 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
7XK5
 
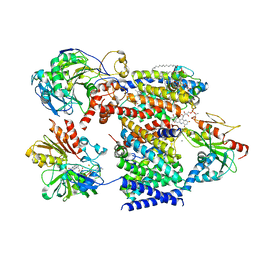 | | Cryo-EM structure of Na+-pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae, state 3 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kishikawa, J, Ishikawa, M, Masuya, T, Murai, M, Barquera, B, Miyoshi, H. | | Deposit date: | 2022-04-19 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Na + -pumping NADH-ubiquinone oxidoreductase from Vibrio cholerae.
Nat Commun, 13, 2022
|
|
