5U8R
 
 | |
9LNP
 
 | | the hUNG bound to DNA product embedding uridine ribonucleotide | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*T*(RP5)P*AP*TP*CP*TP*T)-3'), ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
9LNQ
 
 | | The hUNG bound to DNA product embedding 4primer-OCH3-dU | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(KBC)P*AP*TP*CP*TP*T)-3'), SODIUM ION, ... | | Authors: | Liu, Y, Zhou, C, Zhan, X, Fan, C. | | Deposit date: | 2025-01-21 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uridine Embedded within DNA is Repaired by Uracil DNA Glycosylase via a Mechanism Distinct from That of Ribonuclease H2.
J.Am.Chem.Soc., 147, 2025
|
|
4CZT
 
 | | Crystal structure of the kinase domain of CIPK23 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CBL-INTERACTING SERINE/THREONINE-PROTEIN KINASE 23, SULFATE ION | | Authors: | Chaves-Sanjuan, A, Sanchez-Barrena, M.J, Albert, A. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Regulatory Mechanism of the Plant Cipk Family of Protein Kinases Controlling Ion Homeostasis and Abiotic Stress
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CZU
 
 | |
4D28
 
 | |
9IZN
 
 | | Crystal structure of HKU1A RBD bound to TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Wang, W, Xu, Y, Zhang, S. | | Deposit date: | 2024-08-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of human coronavirus HKU1 receptor binding domain bound to TMPRSS2 receptor
Hlife, 2024
|
|
4MHE
 
 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
7NKT
 
 | | RBD domain of SARS-CoV2 in complex with neutralizing nanobody NM1226 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Ostertag, E, Zocher, G, Stehle, T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Embo Rep., 22, 2021
|
|
8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
8ZE6
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to MjDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-04 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
4RGF
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme soaked with Mn2+ | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2008 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4RGE
 
 | | Crystal structure of the in-line aligned env22 twister ribozyme | | Descriptor: | MAGNESIUM ION, env22 twister ribozyme | | Authors: | Ren, A, Rajashankar, K.R, Simanshu, D, Patel, D. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | In-line alignment and Mg(2+) coordination at the cleavage site of the env22 twister ribozyme.
Nat Commun, 5, 2014
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
5CPR
 
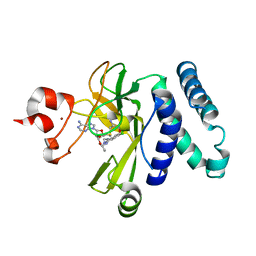 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
4V4W
 
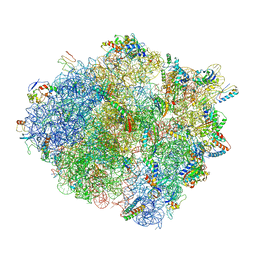 | |
6EW9
 
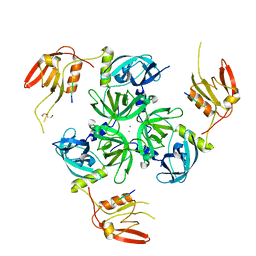 | | CRYSTAL STRUCTURE OF DEGS STRESS SENSOR PROTEASE IN COMPLEX WITH ACTIVATING DNRLGLVYQF PEPTIDE | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DNRLGLVYQF PEPTIDE, ... | | Authors: | Vetter, I.R, Porfetye, A.T, Stege, P. | | Deposit date: | 2017-11-03 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Noncatalytic Lysine Residues from Allosteric Circuits via Covalent Probes.
ACS Chem. Biol., 13, 2018
|
|
4V4V
 
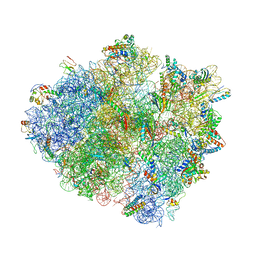 | |
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
6F5O
 
 | |
6F5P
 
 | |
8AN3
 
 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
8AN2
 
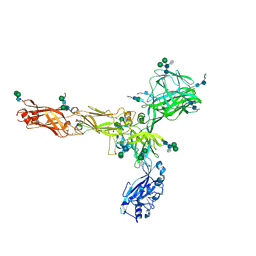 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 10.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
3N57
 
 | |
3N56
 
 | |
