8FG6
 
 | |
3OB6
 
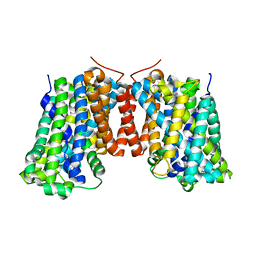 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | Descriptor: | ARGININE, AdiC arginine:agmatine antiporter | | Authors: | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6GVV
 
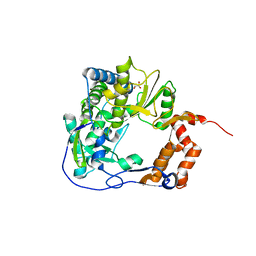 | |
4WYL
 
 | | Mutant K18E of 3D polymerase from Foot-and-Moth Disease Virus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, de la Higuera, I, Domingo, E. | | Deposit date: | 2014-11-17 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
8TDH
 
 | |
4WYW
 
 | | Mutant K20E of 3D polymerase from Foot-and-Mouth Disease Virus | | Descriptor: | ACETATE ION, GLYCEROL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, de la Higuera, I, Domingo, E. | | Deposit date: | 2014-11-18 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
8TDE
 
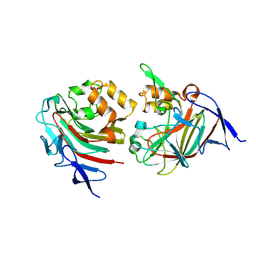 | |
8TDA
 
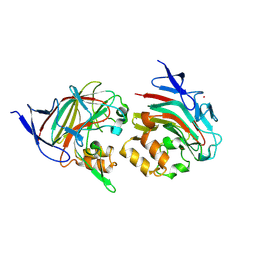 | |
8TDF
 
 | |
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | Descriptor: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TDI
 
 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-03 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TCS
 
 | | Structure of trehalose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, Xylose isomerase-like TIM barrel domain-containing protein, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TCR
 
 | | Structure of glucose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | COBALT (II) ION, MALONATE ION, Sugar phosphate isomerase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8V31
 
 | |
4X2B
 
 | |
5IFE
 
 | | Crystal structure of the human SF3b core complex | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Cretu, C, Dybkov, O, De Laurentiis, E, Will, C.L, Luhrmann, R, Pena, V. | | Deposit date: | 2016-02-25 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations.
Mol.Cell, 64, 2016
|
|
4WZM
 
 | | Mutant K18E of RNA dependent RNA polymerase from Foot-and-Mouth Disease Virus complexed with RNA | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Verdaguer, N, Ferrer-Orta, C, de la Higuera, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
4WZQ
 
 | |
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7Q3E
 
 | | Structure of the mouse CPLANE-RSG1 complex | | Descriptor: | Ciliogenesis and planar polarity effector 2, GUANOSINE-5'-TRIPHOSPHATE, Protein fuzzy homolog, ... | | Authors: | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
7Q3D
 
 | | Structure of the human CPLANE complex | | Descriptor: | Protein fuzzy homolog, Protein inturned, WD repeat-containing and planar cell polarity effector protein fritz homolog | | Authors: | Langousis, G, Cavadini, S, Kempf, G, Matthias, P. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the ciliogenesis-associated CPLANE complex.
Sci Adv, 8, 2022
|
|
6VGQ
 
 | | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, Z-Gly-leu-phe-CH2Cl | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VGK
 
 | | ClpP1P2 complex from M. tuberculosis | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
