5SRB
 
 | |
5SRA
 
 | |
5SPK
 
 | |
5SPL
 
 | |
5SPJ
 
 | |
5SR9
 
 | |
5SPN
 
 | |
5SPM
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00002410346 | | Descriptor: | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPO
 
 | |
5SPP
 
 | |
5SPR
 
 | |
5SPU
 
 | |
5SPT
 
 | |
5SPS
 
 | |
5SPV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with REAL250003774401 | | Descriptor: | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SPQ
 
 | |
5SPW
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00004674769 - (R,S,R) isomer | | Descriptor: | (1R,5S,6R)-3-(7H-purin-6-yl)-3-azabicyclo[3.2.2]nonane-6-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SOJ
 
 | |
5SOQ
 
 | |
5SOI
 
 | |
5HG7
 
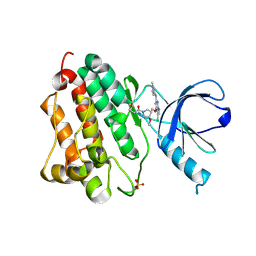 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | Descriptor: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
7UJ7
 
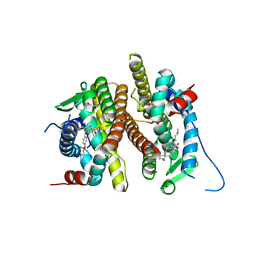 | |
7UJC
 
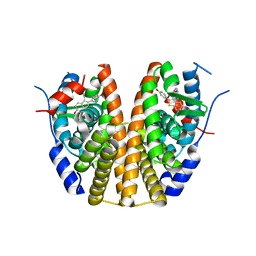 | |
7UJ8
 
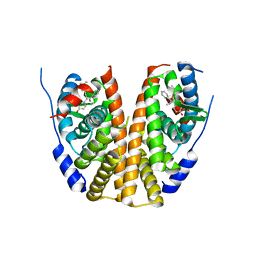 | |
7UJF
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with a Methylated Lasofoxifene Derivative with Selective Estrogen Receptor Degrader Properties | | Descriptor: | (5R,6S)-5-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-6-phenyl-5,6,7,8-tetrahydronaphthalen-2-ol, Estrogen receptor | | Authors: | Hosfield, D.J, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-03-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stereospecific lasofoxifene derivatives reveal the interplay between estrogen receptor alpha stability and antagonistic activity in ESR1 mutant breast cancer cells.
Elife, 11, 2022
|
|
