7DT1
 
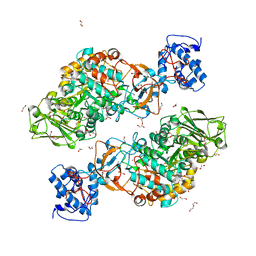 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
5M0K
 
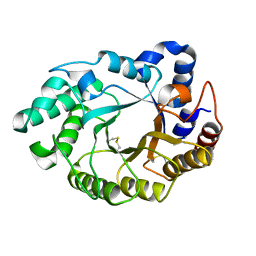 | |
5M6G
 
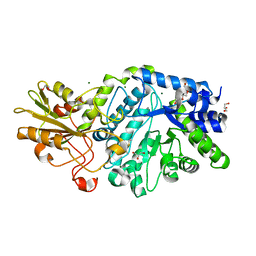 | | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea | | Descriptor: | Beta-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea
To Be Published
|
|
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | Authors: | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | Deposit date: | 2000-05-07 | | Release date: | 2000-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5MRR
 
 | | Crystal structure of L1 protease of Lysobacter sp. XL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of L1 protease of Lysobacter sp. XL1
To Be Published
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
5MRT
 
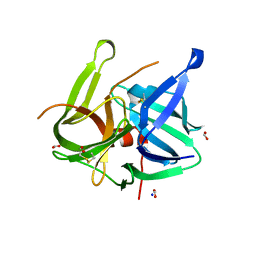 | | Crystal structure of L5 protease Lysobacter sp. XL1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L5 protease Lysobacter sp. XL1
To Be Published
|
|
5MRJ
 
 | | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum | | Descriptor: | Beta-xylanase, SULFATE ION | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum
To Be Published
|
|
5MRS
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF
To Be Published
|
|
7OMT
 
 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7A8V
 
 | | Crystal structure of Polysaccharide monooxygenase from P.verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-08-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94961083 Å) | | Cite: | Crystal structure of Polysaccharide monooxygenase from P.verruculosum
To Be Published
|
|
5L9C
 
 | | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Nemashkalov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinisyn, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose
to be published
|
|
6YON
 
 | | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, SULFATE ION | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum
To Be Published
|
|
1DLG
 
 | | CRYSTAL STRUCTURE OF THE C115S ENTEROBACTER CLOACAE MURA IN THE UN-LIGANDED STATE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE MURA | | Authors: | Schonbrunn, E, Eschenburg, S, Krekel, F, Luger, K, Amrhein, N. | | Deposit date: | 1999-12-09 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA.
Biochemistry, 39, 2000
|
|
1CJS
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 50S RIBOSOMAL PROTEIN L1P | | Authors: | Nevskaya, N, Tishchenko, S, Fedorov, R, Al-Karadaghi, S, Liljas, A, Kraft, A, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 1999-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family.
Structure Fold.Des., 8, 2000
|
|
6T9F
 
 | | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24847341 Å) | | Cite: | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
6T9G
 
 | | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29620671 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
6TPC
 
 | | Crystal structure of Endoglucanase N194A from Penicillium verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, PHOSPHATE ION, ... | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5221895 Å) | | Cite: | Crystal structure of Endoglucanase N194A from Penicillium verruculosum
To Be Published
|
|
5I6S
 
 | | Crystal structure of an endoglucanase from Penicillium verruculosum | | Descriptor: | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, endoglucanase | | Authors: | Nemashklaov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinitsyn, A. | | Deposit date: | 2016-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum
To Be Published
|
|
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CE7
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
1IC6
 
 | | STRUCTURE OF A SERINE PROTEASE PROTEINASE K FROM TRITIRACHIUM ALBUM LIMBER AT 0.98 A RESOLUTION | | Descriptor: | CALCIUM ION, NITRATE ION, PROTEINASE K | | Authors: | Betzel, C, Gourinath, S, Kumar, P, Kaur, P, Perbandt, M, Eschenburg, S, Singh, T.P. | | Deposit date: | 2001-03-30 | | Release date: | 2001-04-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure of a serine protease proteinase K from Tritirachium album limber at 0.98 A resolution.
Biochemistry, 40, 2001
|
|
1F7Y
 
 | | THE CRYSTAL STRUCTURE OF TWO UUCG LOOPS HIGHLIGHTS THE ROLE PLAYED BY 2'-HYDROXYL GROUPS IN ITS UNUSUAL STABILITY | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Nikouline, A, Serganov, A, Tishchenko, S, Nevskaya, N, Garber, M, Ehresmann, B, Ehresmann, C, Nikonov, S, Dumas, P. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of UUCG tetraloop.
J.Mol.Biol., 304, 2000
|
|
1NAW
 
 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
