3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J3U
 
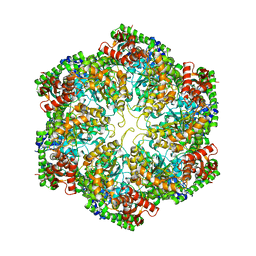 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3JCP
 
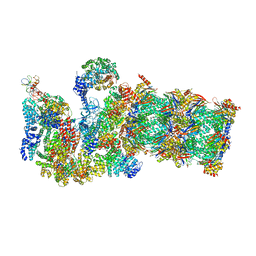 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
6U5M
 
 | |
6U8B
 
 | |
6U6W
 
 | |
6U8O
 
 | |
6U8L
 
 | |
3RLQ
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5- carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1HM1
 
 | | THE SOLUTION NMR STRUCTURE OF A THERMALLY STABLE FAPY ADDUCT OF AFLATOXIN B1 IN AN OLIGODEOXYNUCLEOTIDE DUPLEX REFINED FROM DISTANCE RESTRAINED MOLECULAR DYNAMICS SIMULATED ANNEALING, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA (5'-D(TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3') | | Authors: | Mao, H, Deng, Z, Wang, F, Harris, T.M, Stone, M.P. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR.
Biochemistry, 37, 1998
|
|
8TIF
 
 | | Cryo-EM of mono-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM of mono-pilus from S. islandicus REY15A
To Be Published
|
|
8TIB
 
 | | Cryo-EM of tri-pilus from S. islandicus REY15A | | Descriptor: | DUF973 family protein | | Authors: | Eastep, G.N, Liu, J, Rich-New, S.T, Egelman, E.H, Krupovic, M, Wang, F. | | Deposit date: | 2023-07-19 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM of tri-pilus from S. islandicus REY15A
To Be Published
|
|
3RLR
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RLP
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4H86
 
 | | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | Peroxiredoxin type-2 | | Authors: | Liu, M, Wang, F, Qiu, R, Wu, T, Gu, S, Tang, R, Ji, C. | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of Ahp1 from Saccharomyces cerevisiae in reduced form
To be Published
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
3TLQ
 
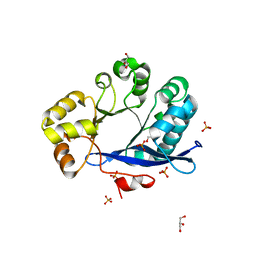 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
6K9N
 
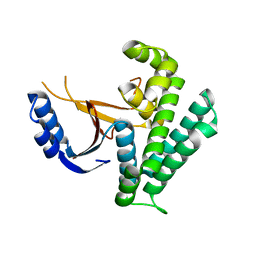 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6KBE
 
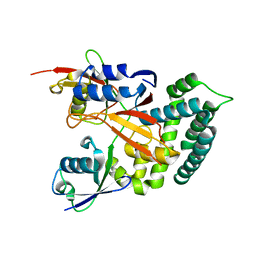 | | Structure of Deubiquitinase | | Descriptor: | Polyubiquitin-C, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
6K9P
 
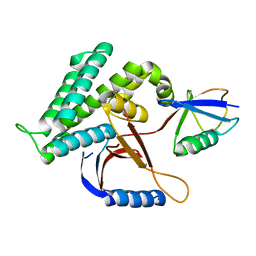 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
4KN8
 
 | | Crystal structure of Bs-TpNPPase | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
4Y2M
 
 | | apo-GolB protein | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Zhao, J, Wang, F. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
