6LBO
 
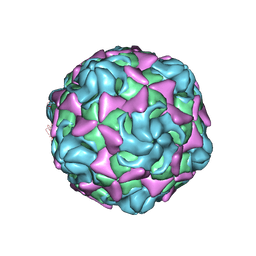 | | Cryo-EM structure of echovirus 11 empty particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LBQ
 
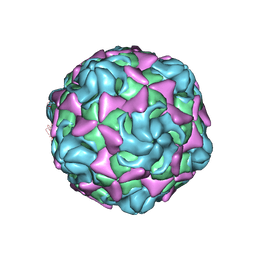 | | Cryo-EM structure of echovirus 11 empty particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LJA
 
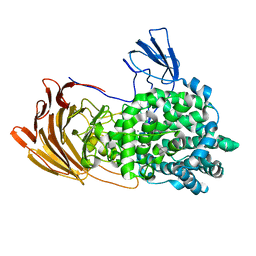 | | Crystal Structure of exoHep from Bacteroides intestinalis DSM 17393 complexed with disaccharide product | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
6KZK
 
 | |
6LA4
 
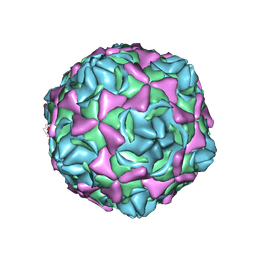 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
8I5N
 
 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8I5M
 
 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
6LA6
 
 | |
6LAP
 
 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LJL
 
 | | Crystal Structure of exoHep-Y390A/H555A complexed with a tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, Heparinase II/III-like protein | | Authors: | Zhang, Q.D, Cao, H.Y, Wei, L, Li, F.C, Zhang, Y.Z. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of exolytic heparinases and their catalytic mechanism and potential application.
Nat Commun, 12, 2021
|
|
4XVC
 
 | |
6MT0
 
 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolodihydropyrrolone inhibitor | | Descriptor: | 3-(1-methylcyclopropyl)-2-[(1-methylcyclopropyl)amino]-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2018-10-18 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a Potent and Selective Pim-1/2 Kinase Inhibitor for Hematological Malignancies.
J. Med. Chem., 62, 2019
|
|
5GZR
 
 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
7XF5
 
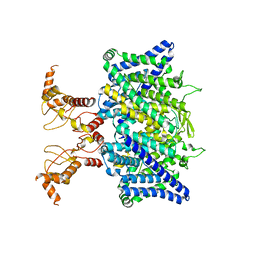 | | Full length human CLC-2 channel in apo state | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-01 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
5E6O
 
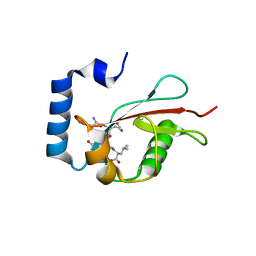 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
7XJA
 
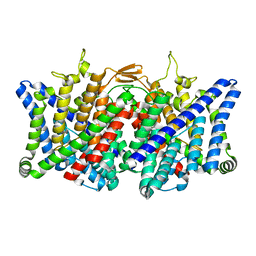 | | TMD masked refine map of human ClC-2 | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
5GMS
 
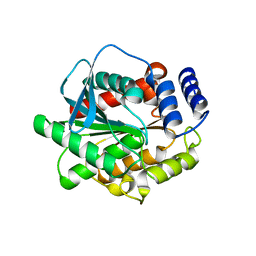 | | Crystal structure of the mutant S202W/I203F of the esterase E40 | | Descriptor: | Esterase | | Authors: | Zhang, Y.-Z, Li, P.-Y. | | Deposit date: | 2016-07-15 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Mechanistic Insights into the Improvement of the Halotolerance of a Marine Microbial Esterase by Increasing Intra- and Interdomain Hydrophobic Interactions.
Appl. Environ. Microbiol., 83, 2017
|
|
5E6N
 
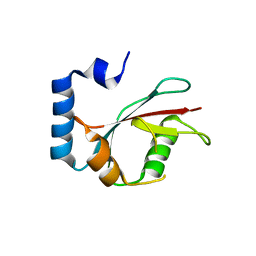 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5EOL
 
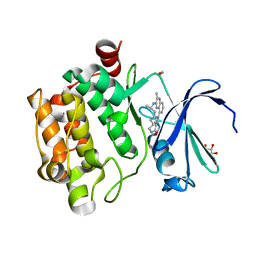 | |
5IPJ
 
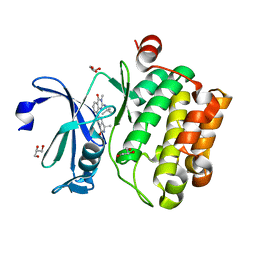 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolopyrrolone inhibitor. | | Descriptor: | 2-(tert-butylamino)-3-methyl-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Quinazolinone-pyrrolopyrrolones as Potent and Orally Bioavailable Pan-Pim Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
8KER
 
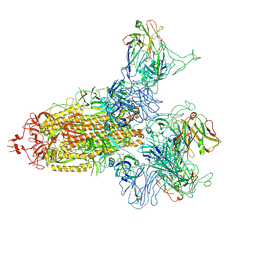 | |
8KDT
 
 | |
8KDS
 
 | |
8KEP
 
 | |
8KEK
 
 | |
