3PP0
 
 | | Crystal Structure of the Kinase domain of Human HER2 (erbB2). | | Descriptor: | 2-{2-[4-({5-chloro-6-[3-(trifluoromethyl)phenoxy]pyridin-3-yl}amino)-5H-pyrrolo[3,2-d]pyrimidin-5-yl]ethoxy}ethanol, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Skene, R.J, Aertgeerts, K, Sogabe, S. | | Deposit date: | 2010-11-23 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analysis of the Mechanism of Inhibition and Allosteric Activation of the Kinase Domain of HER2 Protein.
J.Biol.Chem., 286, 2011
|
|
7F8K
 
 | |
6KAM
 
 | | Crystal structure of FKRP in complex with Ba ion, CDP-ribtol, and sugar acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAL
 
 | | Crystal structure of FKRP in complex with Mg ion and CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAN
 
 | | Crystal structure of FKRP in complex with Ba ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAK
 
 | | Crystal structure of FKRP in complex with Mg ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6KAJ
 
 | | Crystal structure of FKRP in complex with Ba ion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2249 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
5ZF0
 
 | | X-ray Structure of the Electron Transfer Complex between Ferredoxin and Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kubota-Kawai, H, Mutoh, R, Shinmura, K, Setif, P, Nowaczyk, M, Roegner, M, Ikegami, T, Tanaka, T, Kurisu, G. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex
Nat Plants, 4, 2018
|
|
3WUM
 
 | |
3WXT
 
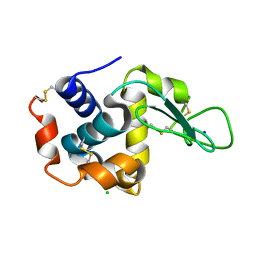 | |
3WXU
 
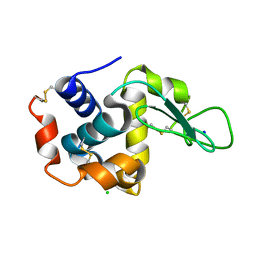 | |
7ZY3
 
 | | Room temperature structure of Archaerhodopsin-3 obtained 110 ns after photoexcitation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kwan, T.O.C, Judge, P.J, Moraes, I, Watts, A, Axford, D, Bada Juarez, J.F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
5WZN
 
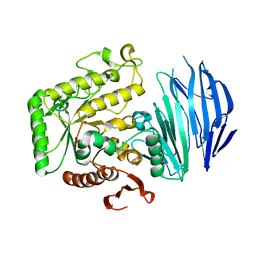 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
9JTQ
 
 | | Crystal structure of the light-driven proton pump heimdallarchaeial rhodopsin HeimdallR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, NITRATE ION, RETINAL, ... | | Authors: | Matsuzaki, Y, Tzlil, G, Del Carmen Marin, M, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2024-10-06 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-harvesting by antenna-containing rhodopsins in pelagic Asgard archaea
To Be Published
|
|
9KDG
 
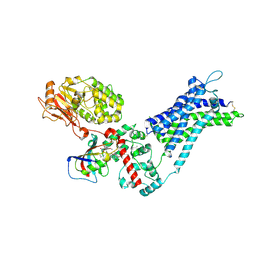 | | CryoEM structure of Calcineurin-fusion Human endothelin receptor type-B in the ligand-free form | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, Calcineurin-fusion endothelin receptor type-B, ... | | Authors: | Shihoya, W, Akasaka, H, Nureki, O. | | Deposit date: | 2024-11-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure of a lasso peptide bound ET B receptor provides insights into the mechanism of GPCR inverse agonism.
Nat Commun, 16, 2025
|
|
9KDF
 
 | | CryoEM structure of Calcineurin-fusion Human endothelin receptor type-B in complex with RES-701-3 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, Calcineurin-fusion endothelin receptor type-B, ... | | Authors: | Shihoya, W, Akasaka, H, Nureki, O. | | Deposit date: | 2024-11-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a lasso peptide bound ET B receptor provides insights into the mechanism of GPCR inverse agonism.
Nat Commun, 16, 2025
|
|
2CZU
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
2CZT
 
 | | lipocalin-type prostaglandin D synthase | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-17 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2ROH
 
 | | The DNA binding domain of RTBP1 | | Descriptor: | Telomere binding protein-1 | | Authors: | Lee, W, Ko, S. | | Deposit date: | 2008-03-22 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of rice telomere binding protein RTBP1
Biochemistry, 48, 2009
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
7YNH
 
 | |
