2LYJ
 
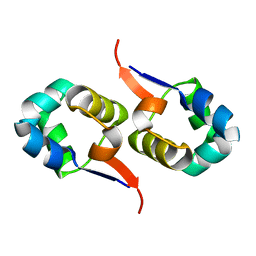 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
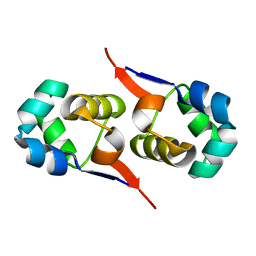 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
3H0T
 
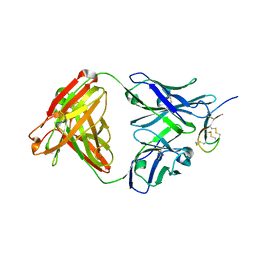 | | Hepcidin-Fab complex | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Syed, R, Li, V. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
2LYS
 
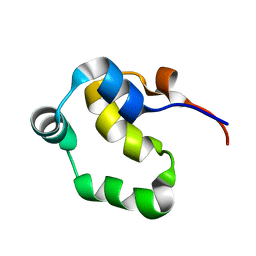 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
4QBQ
 
 | | Crystal structure of DNMT3a ADD domain bound to H3 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone H3, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2014-05-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
To be Published
|
|
2LYL
 
 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
8VB0
 
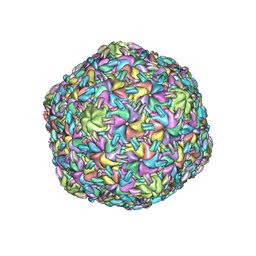 | |
8JZM
 
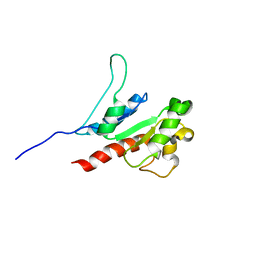 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
3SJE
 
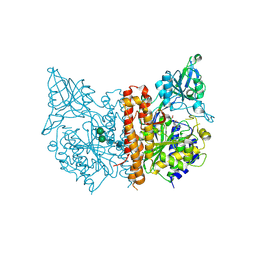 | | X-ray structure of human glutamate carboxypeptidase II (the E424A inactive mutant) in complex with N-acetyl-aspartyl-aminononanoic acid | | Descriptor: | (2S)-2-[(N-acetyl-L-alpha-aspartyl)amino]nonanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
8VBX
 
 | |
8VB4
 
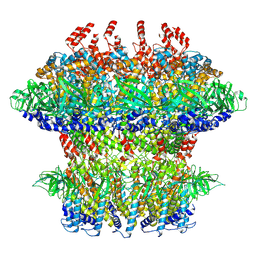 | |
8VB2
 
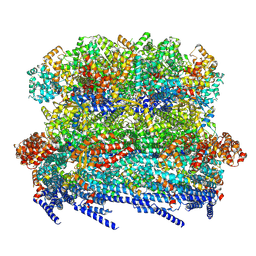 | |
3EZN
 
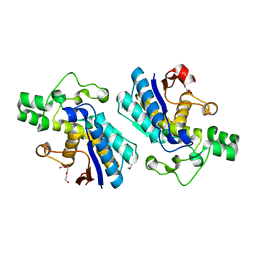 | |
8JPS
 
 | | Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map) | | Descriptor: | Atypical chemokine receptor 1, C-C motif chemokine 7 | | Authors: | Banerjee, R, Khanppnavar, B, Maharana, J, Saha, S, Korkhov, V.M, Shukla, A.K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.
Cell, 187, 2024
|
|
3FDZ
 
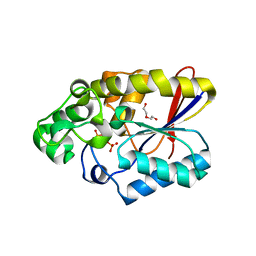 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3FTY
 
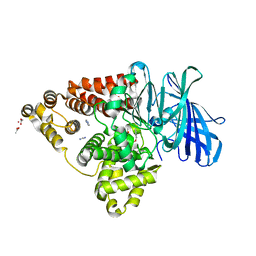 | |
3FUF
 
 | |
3FU0
 
 | |
3FUH
 
 | |
3FU3
 
 | |
3FUK
 
 | |
3FTX
 
 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FUI
 
 | |
