6GV8
 
 | |
1BBD
 
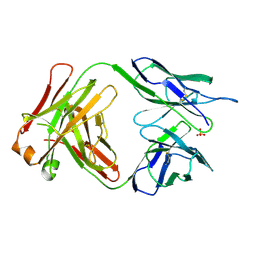 | |
1B1U
 
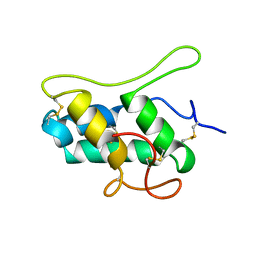 | |
4E1V
 
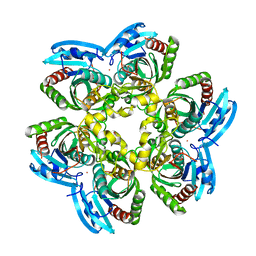 | | X-RAY Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with 5-Fluorouracil at 2.15 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Lashkov, A.A, Sotnichenko, S.E, Prokofev, I.I, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structure of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil and molecular modelling of the complex of 5-fluorouracil with uridine phosphorylase from Vibrio cholerae.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6JQQ
 
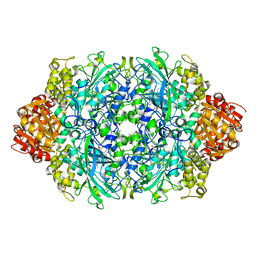 | | KatE H392C from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, J.B, Cho, H.-S. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | KatE H392C from Escherichia coli
To Be Published
|
|
2NUV
 
 | | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with atenolol at 2.25 A resolution | | Descriptor: | 2-(4-(2-HYDROXY-3-(ISOPROPYLAMINO)PROPOXY)PHENYL)ETHANAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-11-10 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with atenolol at 2.25 A resolution
To be Published
|
|
6RCA
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with 2.2'-anhydrouridine at 1.34 A | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-Anhydro-(1-beta-D-ribofuranosyl)uracil, CHLORIDE ION, ... | | Authors: | Prokofev, I.I, Eistrikh-Geller, P.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | X-Ray Structure and Molecular Dynamics Study of Uridine Phosphorylase from Vibrio cholerae in Complex with 2,2'-Anhydrouridine
Crystallography Reports, 2020
|
|
2PX1
 
 | | crystal structure of the complex of bovine lactoferrin C-lobe with Ribose at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Vikram, G, Sinha, M, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | crystal structure of the complex of bovine lactoferrin C-lobe with Ribose at 2.5 A resolution
To be Published
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2HZH
 
 | |
2HN9
 
 | |
2HRD
 
 | |
1FQG
 
 | |
1EGQ
 
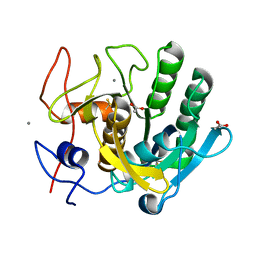 | | ENHANCEMENT OF ENZYME ACTIVITY THROUGH THREE-PHASE PARTITIONING: CRYSTAL STRUCTURE OF A MODIFIED SERINE PROTEINASE AT 1.5 A RESOLUTION | | Descriptor: | ACETIC ACID, CALCIUM ION, PROTEINASE K | | Authors: | Singh, R.K, Gourinath, S, Sharma, S, Ray, I, Gupta, M.N, Singh, T.P. | | Deposit date: | 2000-02-16 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhancement of enzyme activity through three-phase partitioning: crystal structure of a modified serine proteinase at 1.5 A resolution.
Protein Eng., 14, 2001
|
|
1G0Z
 
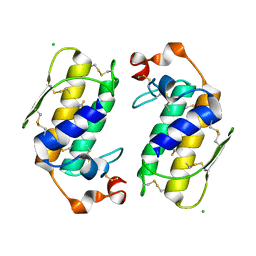 | |
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
1LVL
 
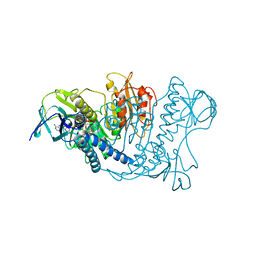 | |
1PC8
 
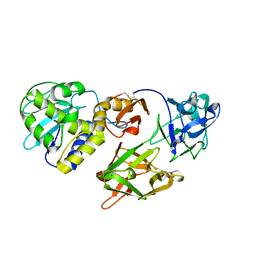 | | Crystal Structure of a novel form of mistletoe lectin from Himalayan Viscum album L. at 3.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Himalayan mistletoe ribosome-inactivating protein, ... | | Authors: | Mishra, V, Ethayathulla, A.S, Paramasivam, M, Singh, G, Yadav, S, Kaur, P, Sharma, R.S, Babu, C.R, Singh, T.P. | | Deposit date: | 2003-05-16 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a novel ribosome-inactivating protein from a hemi-parasitic plant inhabiting the northwestern Himalayas.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TU8
 
 | |
1RDH
 
 | |
1RGB
 
 | | Phospholipase A2 from Vipera ammodytes meridionalis | | Descriptor: | (9E)-OCTADEC-9-ENAMIDE, Phospholipase A2 | | Authors: | Georgieva, D.N. | | Deposit date: | 2003-11-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asp49 phospholipase A(2)-elaidoylamide complex: a new mode of inhibition.
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1SV3
 
 | | Structure of the complex formed between Phospholipase A2 and 4-methoxybenzoic acid at 1.3A resolution. | | Descriptor: | 4-METHOXYBENZOIC ACID, Phospholipase A2, SULFATE ION | | Authors: | Singh, N, Prahathees, E, Jabeen, T, Pal, A, Ethayathulla, A.S, Prem kumar, R, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-27 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding
Proteins, 64, 2006
|
|
1TU7
 
 | |
