7QO7
 
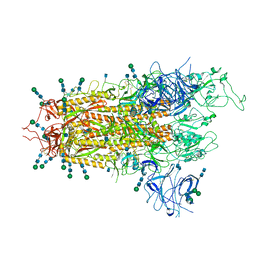 | | SARS-CoV-2 S Omicron Spike B.1.1.529 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Beckert, B, Nazarov, S, Pojer, F, Myasnikov, A, Stahlberg, H, Trono, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion
Biorxiv, 2021
|
|
7SOF
 
 | |
7SOC
 
 | |
1U9C
 
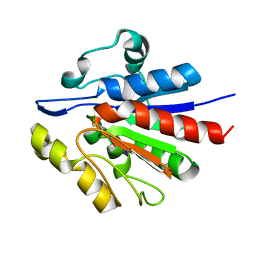 | | Crystallographic structure of APC35852 | | Descriptor: | APC35852 | | Authors: | Borek, D, Chen, Y, Shao, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of DJI superfamily
To be Published
|
|
7SOA
 
 | |
9GC4
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[3-(3-chloranyl-6-fluoranyl-pyridin-2-yl)oxyphenyl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GC6
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[5-fluoranyl-4-(2-fluorophenyl)pyridin-2-yl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GDV
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[2-[2-(dimethylamino)ethyl-methyl-amino]-4-methoxy-5-[[4-(1-methylindol-3-yl)pyrimidin-2-yl]amino]phenyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-06 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9GC5
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[5-(1,3-benzoxazol-4-yl)-2,4-bis(fluoranyl)phenyl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
7Q20
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) in complex with blood group A trisaccharide | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Ruminococcus gnavus endogalactosidase GH98, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7AXF
 
 | |
1TY4
 
 | | Crystal structure of a CED-9/EGL-1 complex | | Descriptor: | Apoptosis regulator ced-9, EGg Laying defective EGL-1, programmed cell death activator | | Authors: | Yan, N, Gu, L, Kokel, D, Xue, D, Shi, Y. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4
Mol.Cell, 15, 2004
|
|
8V3U
 
 | | ACAD11 D220A with 4-hydroxyvaleryl-CoA | | Descriptor: | Acyl-Coenzyme A dehydrogenase family, member 11, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, J, Bartlett, A, Rashan, E, Yuan, P, Pagliarini, D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism.
Nat.Struct.Mol.Biol., 2025
|
|
8V3V
 
 | | ACAD11 D753N with 4-phosphovaleryl-CoA | | Descriptor: | Acyl-Coenzyme A dehydrogenase family, member 11, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, J, Bartlett, A, Rashan, E, Yuan, P, Pagliarini, D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism.
Nat.Struct.Mol.Biol., 2025
|
|
1U2C
 
 | |
9GA4
 
 | | MtUvrA2UvrB2 bound to damaged oligonucleotide | | Descriptor: | DNA, UvrABC system protein A, UvrABC system protein B, ... | | Authors: | Genta, M, Capelli, R, Ferrara, G, Rizzi, M, Rossi, F, Jeruzalmi, D, Bolognesi, M, Chaves-Sanjuan, A, Miggiano, R. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic understanding of UvrA damage detection and lesion hand-off to UvrB in Nucleotide Excision Repair.
Nat Commun, 16, 2025
|
|
9FL7
 
 | | Cryo-EM structure of human AK2 bound to reduced human AIFM1 (residues 102-613), class 3 | | Descriptor: | Adenylate kinase 2, mitochondrial, Apoptosis-inducing factor 1, ... | | Authors: | Lauer, S.M, Spahn, C.M.T, Schwefel, D. | | Deposit date: | 2024-06-04 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | An NADH-controlled gatekeeper of ATP synthase.
Mol.Cell, 85, 2025
|
|
7ZFF
 
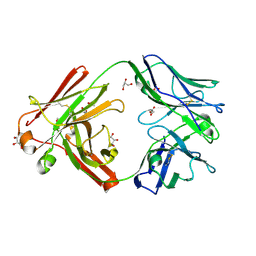 | | Omi-42 Fab | | Descriptor: | GLYCEROL, Omi-42 Heavy chain, Omi-42 light chain | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF6
 
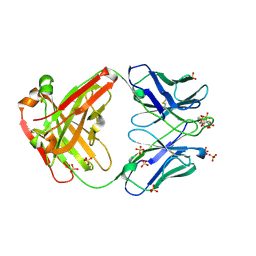 | | Omi-12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF8
 
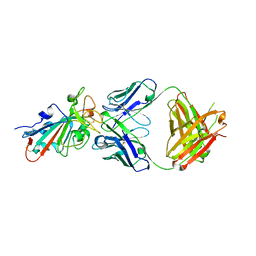 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFE
 
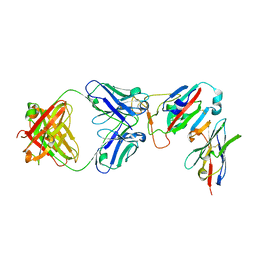 | | SARS-CoV-2 Omicron RBD in complex with Omi-32 Fab and nanobody C1 | | Descriptor: | Nanobody C1, Omi-32 heavy chain, Omi-32 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7AMN
 
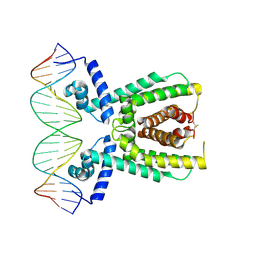 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7ZFD
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-25 Fab | | Descriptor: | Omi-25 heavy chain, Omi-25 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7LMX
 
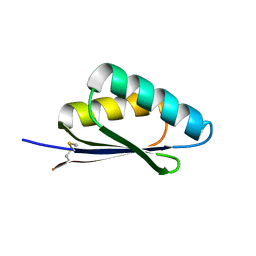 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7ZF3
 
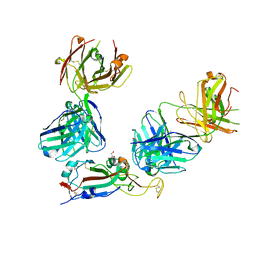 | | SARS-CoV-2 Omicron RBD in complex with Omi-3 and EY6A Fabs | | Descriptor: | EY6A heavy chain, EY6A light chain, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
