2LKM
 
 | |
2M0X
 
 | |
7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XFR
 
 | |
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
6JPJ
 
 | | Crystal structure of FGF401 in complex of FGFR4 | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-7-methanoyl-6-[(4-methyl-2-oxidanylidene-piperazin-1-yl)methyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide, SULFATE ION | | Authors: | Zhou, Z, Chen, X, Chen, Y. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Characterization of FGF401 as a reversible covalent inhibitor of fibroblast growth factor receptor 4.
Chem.Commun.(Camb.), 55, 2019
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
7X7R
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
8WJD
 
 | | Structure of SptTad2 complexed with cGG | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SptTad2 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
8WJE
 
 | | Structure of apo-SPO1 Tad2 | | Descriptor: | Gp34.65 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
8WJC
 
 | | Structure of AcrIIA7 complexed with 3'3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Single phage proteins sequester signals from TIR and CGAS-like enzymes
Nature, 2024
|
|
6JO8
 
 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JJU
 
 | | Structure of Ca2+ ATPase | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Inoue, M, Sakuta, N, Watanabe, S, Inaba, K. | | Deposit date: | 2019-02-27 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Sarco/Endoplasmic Reticulum Ca2+-ATPase 2b Regulation via Transmembrane Helix Interplay.
Cell Rep, 27, 2019
|
|
7V8G
 
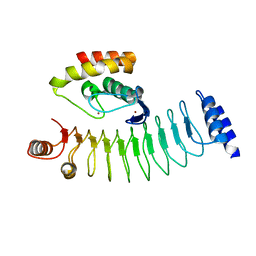 | | Crystal structure of HOIP RING1 domain bound to IpaH1.4 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31, RING-type E3 ubiquitin transferase, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8E
 
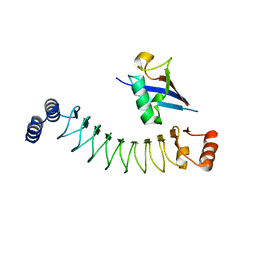 | | Crystal structure of IpaH1.4 LRR domain bound to HOIL-1L UBL domain. | | Descriptor: | RING-type E3 ubiquitin transferase, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8F
 
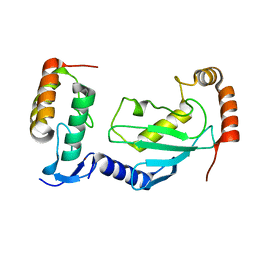 | | Crystal structure of UBE2L3 bound to HOIP RING1 domain. | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin-conjugating enzyme E2 L3, ZINC ION | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7V8H
 
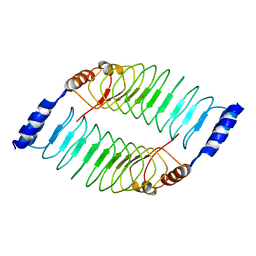 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7EJ1
 
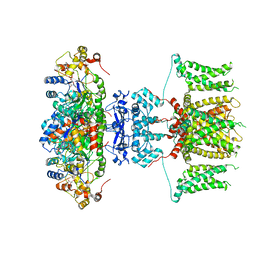 | | human voltage-gated potassium channel KV1.3 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
7EJ2
 
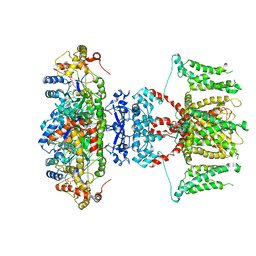 | | human voltage-gated potassium channel KV1.3 H451N mutant | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
8Y1B
 
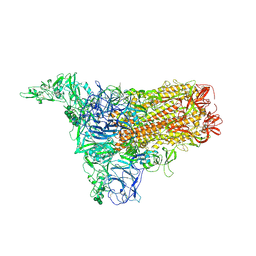 | | 1up-2 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1H
 
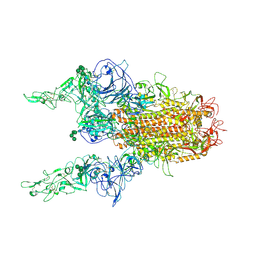 | | The 2up formation of the HKU1-B S protein in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
