2M1U
 
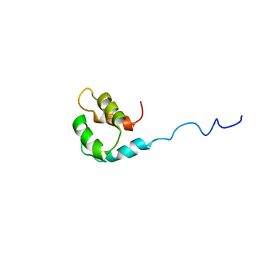 | | Solution structure of the small dictyostelium discoideium myosin light chain mlcb provides insights into iq-motif recognition of class i myosin myo1b | | Descriptor: | myosin light chain MlcB | | Authors: | Liburd, J, Chitayat, S, Crawley, S.W, Denis, C.M, Cote, G.P, Smith, S.P. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the small Dictyostelium discoideum myosin light chain MlcB provides insights into MyoB IQ motif recognition.
J.Biol.Chem., 289, 2014
|
|
2M8U
 
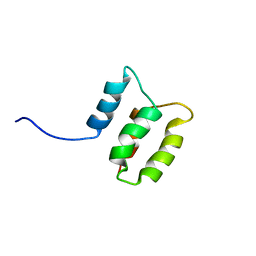 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2MH0
 
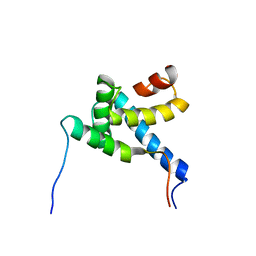 | |
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
2H0B
 
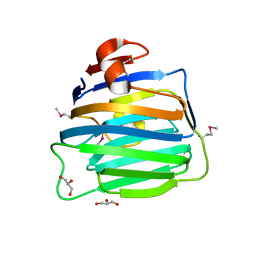 | | Crystal Structure of the second LNS/LG domain from Neurexin 1 alpha | | Descriptor: | CALCIUM ION, GLYCEROL, Neurexin-1-alpha | | Authors: | Sheckler, L.R, Henry, L, Sugita, S, Sudhof, T.C, Rudenko, G. | | Deposit date: | 2006-05-14 | | Release date: | 2006-06-20 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Second LNS/LG Domain from Neurexin 1{alpha}: Ca2+ binding and the effects of alternative splicing
J.Biol.Chem., 281, 2006
|
|
1GC6
 
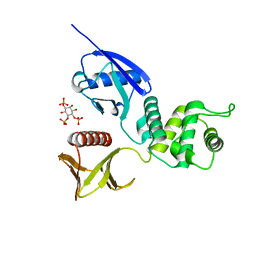 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1GC7
 
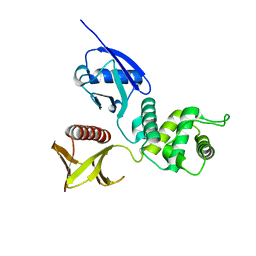 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN | | Descriptor: | RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
2CWN
 
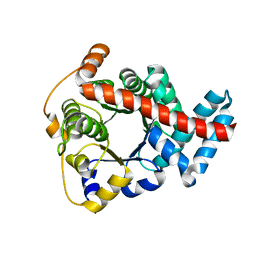 | | Crystal structure of mouse transaldolase | | Descriptor: | Transaldolase | | Authors: | Handa, N, Arai, R, Nishino, A, Uchikubo, T, Takemoto, C, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
2CY4
 
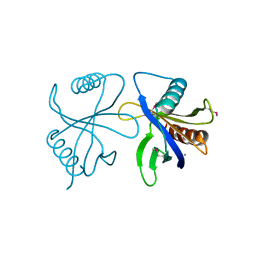 | | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-1 crystal) | | Descriptor: | CALCIUM ION, epidermal growth factor receptor pathway substrate 8-like protein 1 | | Authors: | Mizohata, E, Hamana, H, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-1 crystal)
To be Published
|
|
2CZ2
 
 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal) | | Descriptor: | GLUTATHIONE, GLYCEROL, Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-1 crystal)
To be Published
|
|
2CY5
 
 | | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal) | | Descriptor: | CALCIUM ION, epidermal growth factor receptor pathway substrate 8-like protein 1 | | Authors: | Mizohata, E, Hamana, H, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphotyrosine binding (PTB) domain of epidermal growth factor receptor pathway substrate-8 (EPS8) related protein 1 from Mus musculus (form-2 crystal)
To be Published
|
|
2CZ3
 
 | | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal) | | Descriptor: | Maleylacetoacetate isomerase | | Authors: | Mizohata, E, Morita, S, Kinoshita, Y, Nagano, K, Uda, H, Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-10 | | Release date: | 2006-01-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutathione transferase zeta 1-1 (maleylacetoacetate isomerase) from Mus musculus (form-2 crystal)
To be Published
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | Descriptor: | Claudin-19, Heat-labile enterotoxin B chain | | Authors: | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
1WP1
 
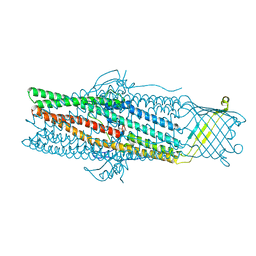 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
6AID
 
 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
1J8K
 
 | | NMR STRUCTURE OF THE FIBRONECTIN EDA DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Niimi, T, Osawa, M, Yamaji, N, Yasunaga, K, Sakashita, H, Mase, T, Tanaka, A, Fujita, S. | | Deposit date: | 2001-05-22 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human fibronectin EDA.
J.Biomol.NMR, 21, 2001
|
|
3VKM
 
 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3AZ4
 
 | | Crystal structure of Co/O-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ5
 
 | | Crystal structure of Pt/O-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ6
 
 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
3AZ7
 
 | | Crystal structure of Pt/T-HEWL | | Descriptor: | Lysozyme C, PLATINUM (II) ION, SODIUM ION | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
2D8N
 
 | | Crystal structure of human recoverin at 2.2 A resolution | | Descriptor: | Recoverin | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-07 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human recoverin at 2.2 A resolution
To be Published
|
|
3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | Descriptor: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
