5YKO
 
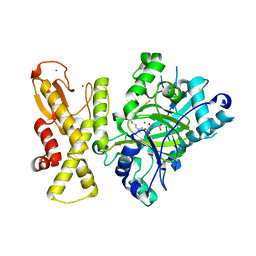 | |
5YKN
 
 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
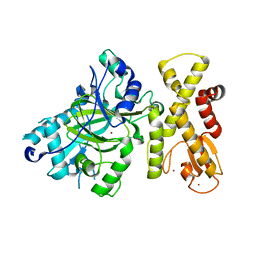
7CWQ
 
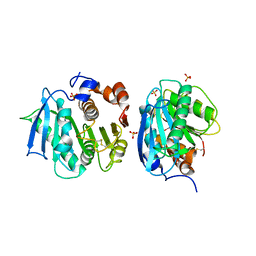 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7CVQ
 
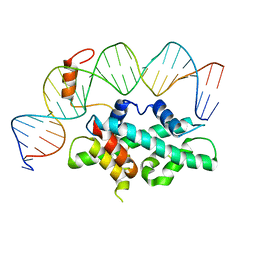 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
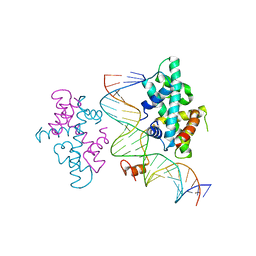 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CY0
 
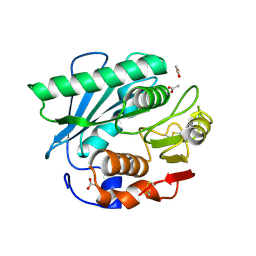 | | Crystal structure of S185H mutant PET hydrolase from Ideonella sakaiensis | | Descriptor: | ACETIC ACID, Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7F5X
 
 | | GK domain of Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-L-GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5V
 
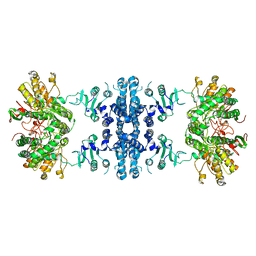 | | Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5U
 
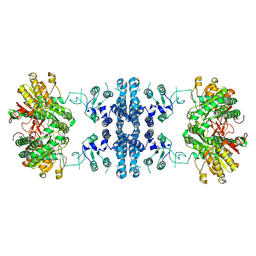 | |
