2F9Q
 
 | | Crystal Structure of Human Cytochrome P450 2D6 | | Descriptor: | Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Rowland, P. | | Deposit date: | 2005-12-06 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6
J.Biol.Chem., 281, 2006
|
|
2FYY
 
 | |
1RWS
 
 | |
2FZ3
 
 | |
1SF0
 
 | |
1LEI
 
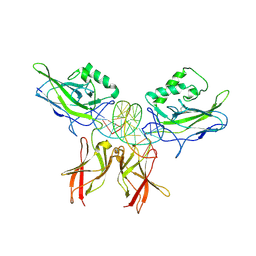 | | The kB DNA sequence from the HLV-LTR functions as an allosteric regulator of HIV transcription | | Descriptor: | 5'-D(*CP*TP*CP*AP*GP*GP*GP*AP*AP*AP*GP*TP*AP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*5ITP*AP*CP*5ITP*5ITP*5ITP*CP*CP*CP*TP*GP*AP*G)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Chen-Park, F, Huang, D.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The kB DNA sequence from the HIV Long Terminal Repeat functions as an allosteric regulator of HIV transcription
J.Biol.Chem., 277, 2002
|
|
3LM5
 
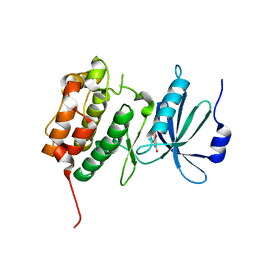 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Serine/threonine-protein kinase 17B | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, Carpenter, L, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
1NNQ
 
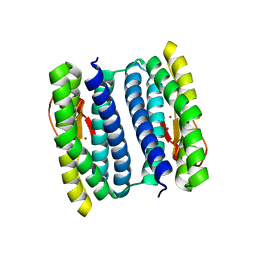 | | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | | Descriptor: | Rubrerythrin, ZINC ION | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin
Proteins, 57, 2004
|
|
1I2Y
 
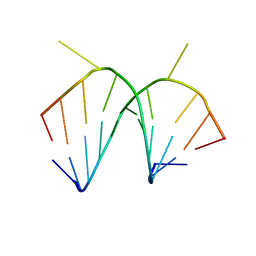 | | 1.66 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
1I2X
 
 | | 2.4 A STRUCTURE OF A-DUPLEX WITH BULGED ADENOSINE, SPERMIDINE FORM | | Descriptor: | DNA/RNA (5'-R(*GP*CP*G)-D(P*AP*TP*AP*T)-R(P*AP*CP*GP*U)-3'), SPERMIDINE | | Authors: | Tereshko, V, Wallace, S, Usman, N, Wincott, F, Egli, M. | | Deposit date: | 2001-02-12 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic observation of "in-line" and "adjacent" conformations in a bulged self-cleaving RNA/DNA hybrid.
RNA, 7, 2001
|
|
2P3J
 
 | |
2P3I
 
 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3K
 
 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2N4P
 
 | | Solution structure of the n-terminal domain of tdp-43 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F, Buratti, E, Laurents, D.V. | | Deposit date: | 2015-06-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The TDP-43 N-terminal domain structure at high resolution.
Febs J., 283, 2016
|
|
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
4XYJ
 
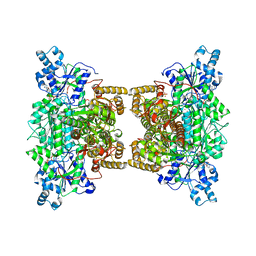 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
3I1C
 
 | |
3G4M
 
 | |
3GES
 
 | |
4XYK
 
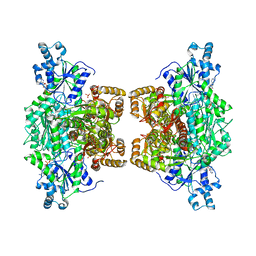 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
4YMC
 
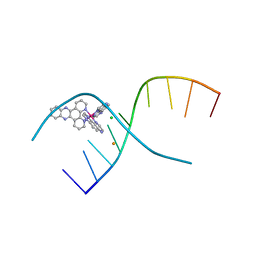 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(CCGGATCCGG)2 | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*GP*AP*TP*CP*CP*GP*G)-3'), ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Reversal of a single base pair step controls guanine photo-oxidation by an intercalating Ru(II) dipyridophenazine complex.
To Be Published
|
|
3GER
 
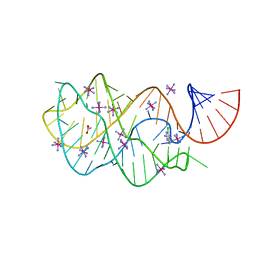 | | Guanine riboswitch bound to 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Gilbert, S.D, Batey, R.T. | | Deposit date: | 2009-02-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Adaptive ligand binding by the purine riboswitch in the recognition of Guanine and adenine analogs
Structure, 17, 2009
|
|
3GAO
 
 | |
3GOG
 
 | |
3U26
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR48 | | Descriptor: | PF00702 domain protein | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
