2WAD
 
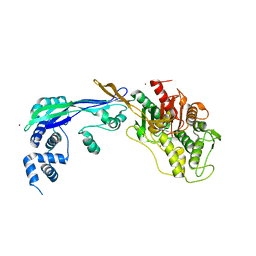 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAF
 
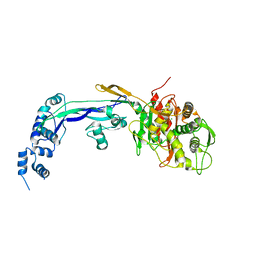 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2V2F
 
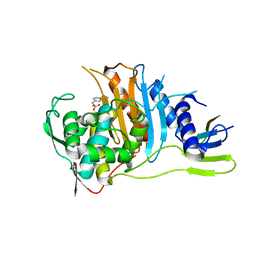 | | Crystal structure of PBP1a from drug-resistant strain 5204 from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BARIUM ION, PENICILLIN BINDING PROTEIN 1A | | Authors: | Job, V, Carapito, R, Vernet, T, Dideberg, O, Dessen, A, Zapun, A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Common Alterations in Pbp1A from Resistant Streptococcus Pneumoniae Decrease its Reactivity Toward {Beta}-Lactams: Structural Insights.
J.Biol.Chem., 283, 2008
|
|
4BLM
 
 | |
4IB5
 
 | | Structure of human protein kinase CK2 catalytic subunit in complex with a CK2beta-competitive cyclic peptide | | Descriptor: | CHLORIDE ION, CK2beta-derived cyclic peptide, Casein kinase II subunit alpha, ... | | Authors: | Raaf, J, Guerra, B, Neundorf, I, Bopp, B, Issinger, O.-G, Jose, J, Pietsch, M, Niefind, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-03-20 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First structure of protein kinase CK2 catalytic subunit with an effective CK2 beta-competitive ligand
Acs Chem.Biol., 8, 2013
|
|
7BN2
 
 | |
7BN3
 
 | |
7BN1
 
 | |
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
3PTE
 
 | |
