8Y6D
 
 | | Norovirus GII.10 P domain and 2'-FL (tablet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GII.10 norovirus P domain in complex with 2'-FL (tablet), ... | | Authors: | Hansman, G, Tame, J.R.H, Kher, G, Pancera, M. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Norovirus GII.10 P domain and 2'-FL (tablet)
To Be Published
|
|
8A7X
 
 | | NaK C-DI F92A mutant soaked in Cs+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CESIUM ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7ZPZ
 
 | | Crystal structure of Pizza6-TSR-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSR-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZQ2
 
 | | Crystal structure of Pizza6-RSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-RSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZQG
 
 | | Crystal structure of Pizza6-KSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-KSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZPW
 
 | | Crystal structure of Pizza6-TSK-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSK-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface.
To be published
|
|
7ZPH
 
 | | Crystal structure of Pizza6-TNK-RSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNK-RSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
5MLN
 
 | |
8AYQ
 
 | | NaK C-DI mutant with Rb+ and Ca2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
8AYP
 
 | | NaK C-DI mutant with Rb+ and Ba2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BARIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
5NBT
 
 | | Apo structure of p60N/p80C katanin | | Descriptor: | Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | Authors: | Jiang, K, Rezabkova, L, Hua, S, Liu, Q, Capitani, G, Altelaar, A.F.M, Heck, A.J.R, Kammerer, R.A, Steinmetz, M.O, Akhmanova, A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
5Z01
 
 | | Native Escherichia coli L,D-carboxypeptidase A (LdcA) | | Descriptor: | MAGNESIUM ION, Murein tetrapeptide carboxypeptidase | | Authors: | Meyer, K, Tame, J.R.H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure and oligomeric form of Escherichia colil,d-carboxypeptidase A.
Biochem. Biophys. Res. Commun., 499, 2018
|
|
5NMR
 
 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
8A35
 
 | | NaK C-DI mutant with Rb+ and Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, RUBIDIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-06-07 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
5Z03
 
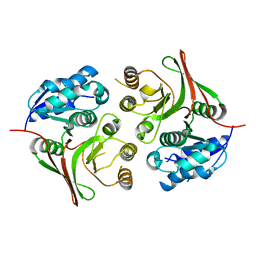 | |
8CMY
 
 | |
5XG5
 
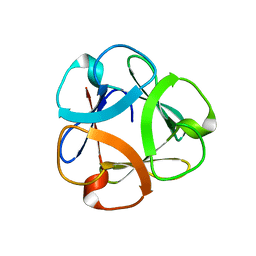 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
8TQP
 
 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-{(1R)-2-(3,5-difluorophenyl)-1-[3-(4-methoxyphenyl)-4-oxo-3,4-dihydroquinazolin-2-yl]ethyl}acetamide, Gag polyprotein | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
8TOV
 
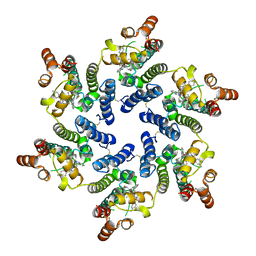 | | HIV-CA Disulfide linked Hexamer bound to Quinazolin-4-one Scaffold inhibitor | | Descriptor: | 2-[4-(4-aminobenzene-1-sulfonyl)-2-oxopiperazin-1-yl]-N-[(1R)-2-(3,5-difluorophenyl)-1-{3-[4-(morpholine-4-sulfonyl)phenyl]-4-oxo-3,4-dihydroquinazolin-2-yl}ethyl]acetamide, Matrix protein p17 | | Authors: | Goldstone, D.C, Barnett, M.J, Taka, J.R.H. | | Deposit date: | 2023-08-04 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, Crystallographic Studies, and Mechanistic Investigations of Novel Phenylalanine Derivatives Bearing a Quinazolin-4-one Scaffold as Potent HIV Capsid Modulators.
J.Med.Chem., 66, 2023
|
|
7MS2
 
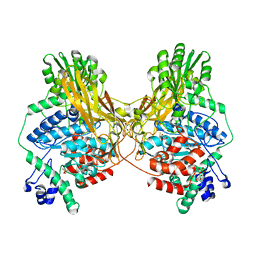 | |
8OJG
 
 | | Structure of the MlaCD complex (2:6 stoichiometry) | | Descriptor: | ABC transporter substrate-binding protein, ABC-type organic solvent transporter | | Authors: | Wotherspoon, P, Bui, S, Sridhar, P, Bergeron, J.R.C, Knowles, T.J. | | Deposit date: | 2023-03-24 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | The structure of the MlaC-MlaD complex reveals novel insights into periplasmic phospholipid transport processes
To Be Published
|
|
8OJ4
 
 | | Structure of the MlaCD complex (1:6 stoichiometry) | | Descriptor: | ABC transporter substrate-binding protein, ABC-type organic solvent transporter | | Authors: | Wotherspoon, P, Bui, S, Sridhar, P, Bergeron, J.R.C, Knowles, T.J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | The structure of the MlaC-MlaD complex reveals novel insights into periplasmic phospholipid transport processes
To Be Published
|
|
5CHB
 
 | | Crystal structure of nvPizza2-S16H58 coordinating a CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
