7AWU
 
 | | Structure of SARS-CoV-2 Main Protease bound to LSN2463359. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | Descriptor: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AOL
 
 | | Structure of SARS-CoV-2 Main Protease bound to Climbazole | | Descriptor: | (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AXO
 
 | | Structure of SARS-CoV-2 Main Protease bound to AR-42. | | Descriptor: | 3C-like proteinase, AR-42, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWW
 
 | | Structure of SARS-CoV-2 Main Protease bound to Clonidine | | Descriptor: | 2,6-DICHLORO-N-IMIDAZOLIDIN-2-YLIDENEANILINE, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AQI
 
 | | Structure of SARS-CoV-2 Main Protease bound to Ifenprodil | | Descriptor: | 3C-like proteinase, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR5
 
 | | Structure of apo SARS-CoV-2 Main Protease with small beta angle, space group C2. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AF0
 
 | | Structure of SARS-CoV-2 Main Protease bound to Ipidacrine. | | Descriptor: | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7APH
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7VQI
 
 | |
6LTG
 
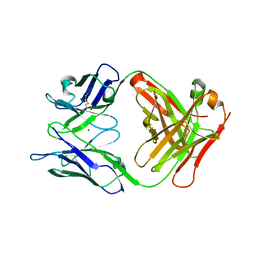 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
5UNG
 
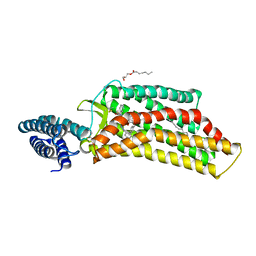 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
7Y52
 
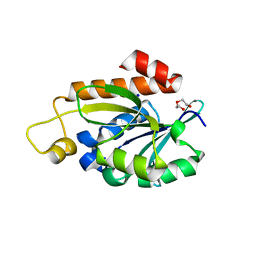 | | Crystal structure of peptidyl-tRNA hydrolase from Enterococcus faecium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Pandey, R, Zohib, M, Mundra, S, Pal, R.K, Arora, A. | | Deposit date: | 2022-06-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of structure of peptidyl-tRNA hydrolase from Enterococcus faecium and its inhibition by a pyrrolinone compound.
Int.J.Biol.Macromol., 275, 2024
|
|
7ZBP
 
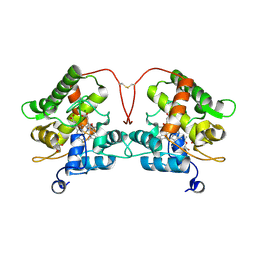 | | Unspecific peroxygenase from Marasmius rotula | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
7ZCL
 
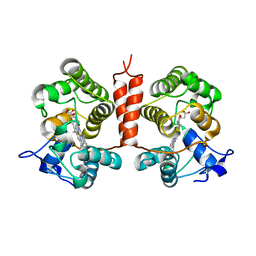 | | Unspecific peroxygenase from Collariella virescens | | Descriptor: | Collariella virescens UPO, HEME C, MAGNESIUM ION | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
6ME3
 
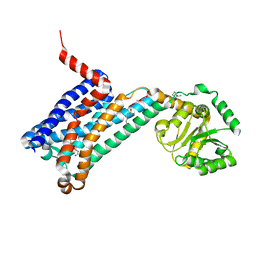 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
7Z28
 
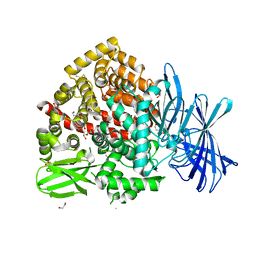 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
6ME5
 
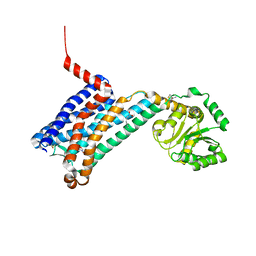 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
6AL2
 
 | | Crystal structure of E. coli YidC at 2.8 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Membrane protein insertase YidC | | Authors: | Tanaka, Y, Tsukazaki, T, Izumioka, A, Hamid, A.A, Fujii, A. | | Deposit date: | 2018-09-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-angstrom crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
7ZE0
 
 | | Solution structure of the PulM C-terminal domain from Klebsiella oxytoca | | Descriptor: | Type II secretion system protein M | | Authors: | Lopez-Castilla, A, Bardiaux, B, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
7Z6G
 
 | |
7Z6J
 
 | |
7Z6D
 
 | |
6MHM
 
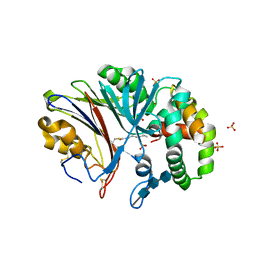 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dementiev, A, Joachimiak, A, Doan, N. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
5P5B
 
 | |
