1VB9
 
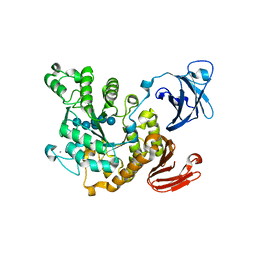 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-amylase II | | Authors: | Mizuno, M, Tonozuka, T, Uechi, A, Ohtaki, A, Ichikawa, K, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-02-25 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) complexed with transglycosylated product
EUR.J.BIOCHEM., 271, 2004
|
|
1ULV
 
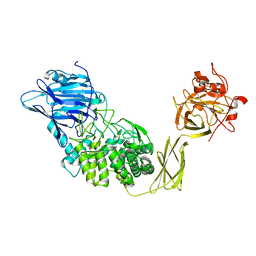 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1SOU
 
 | | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46 | | Descriptor: | 5,10-methenyltetrahydrofolate synthetase | | Authors: | Cort, J.R, Chiang, Y, Acton, T, Wu, M, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46
To be Published
|
|
1T08
 
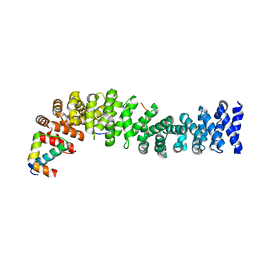 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
1VFO
 
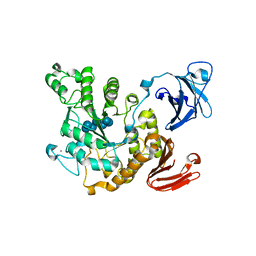 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/beta-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFM
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/alpha-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
2IVW
 
 | | The solution structure of a domain from the Neisseria meningitidis PilP pilot protein. | | Descriptor: | PILP PILOT PROTEIN | | Authors: | Golovanov, A.P, Balasingham, S, Tzitzilonis, C, Goult, B.T, Lian, L.-Y, Homberset, H, Tonjum, T, Derrick, J.P. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold.
J. Mol. Biol., 364, 2006
|
|
2BCO
 
 | | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14 | | Descriptor: | Succinylglutamate desuccinylase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Benach, J, Zhou, W, Acton, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-19 | | Release date: | 2005-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14
To be Published
|
|
3G5V
 
 | | Antibodies Specifically Targeting a Locally Misfolded Region of Tumor Associated EGFR | | Descriptor: | 806 light chain, 808 heavy chain, ACETATE ION, ... | | Authors: | Garrett, T.P.J, Burgess, A.W, Huyton, T, Xu, Y. | | Deposit date: | 2009-02-05 | | Release date: | 2010-02-09 | | Last modified: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Antibodies specifically targeting a locally misfolded region of tumor associated EGFR
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2F6K
 
 | | Crystal Structure of Amidohydrorolase II; Northeast Structural Genomics Target LpR24 | | Descriptor: | MANGANESE (II) ION, metal-dependent hydrolase | | Authors: | Das, K, Xiao, R, Acton, T, Ma, L, Arnold, E, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-29 | | Release date: | 2006-01-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of ACMDS from Lactobacillus plantarum
To be Published
|
|
2D0H
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2D0F
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N complexed with P2, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2BN8
 
 | | Solution Structure and interactions of the E .coli Cell Division Activator Protein CedA | | Descriptor: | CELL DIVISION ACTIVATOR CEDA | | Authors: | Chen, H.A, Simpson, P, Huyton, T, Roper, D, Matthews, S. | | Deposit date: | 2005-03-22 | | Release date: | 2006-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Interactions of the Escherichia Coli Cell Division Activator Protein Ceda.
Biochemistry, 44, 2005
|
|
2D2O
 
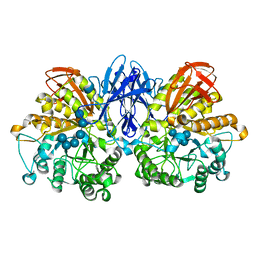 | | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft | | Descriptor: | CALCIUM ION, Neopullulanase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ohtaki, A, Mizuno, M, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-09-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complex of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with maltohexaose demonstrates the important role of aromatic residues at the reducing end of the substrate binding cleft
Carbohydr.Res., 341, 2006
|
|
2B8H
 
 | | A/NWS/whale/Maine/1/84 (H1N9) reassortant influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Smith, B.J, Platis, D, Cox, M.M.J, Huyton, T, Joosten, R.P, McKimm-Breschkin, J.L, Zhang, J.-G, Luo, C.S, Lou, M.-Z, Garrett, T.P.J, Labrou, N.E. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2D0G
 
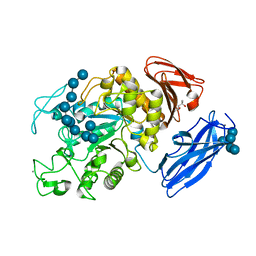 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2EW0
 
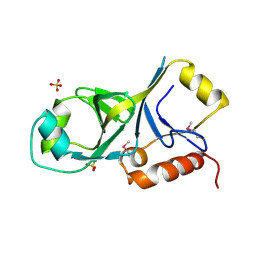 | | X-ray Crystal Structure of Protein Q6FF54 from Acinetobacter sp. ADP1. Northeast Structural Genomics Consortium Target AsR1. | | Descriptor: | SULFATE ION, hypothetical protein ACIAD0353 | | Authors: | Kuzin, A.P, Abashidze, M, Vorobiev, S.M, Forouhar, F, Acton, T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel x-ray structure of hypothetical protein Q6FF54 at the resolution 1.4 A. Northeast Structural Genomics target AsR1.
TO BE PUBLISHED
|
|
1WZK
 
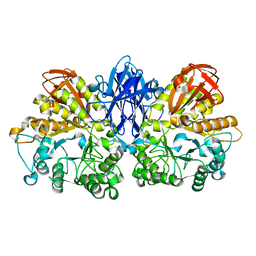 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1IZJ
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1 mutant enzyme f313a | | Descriptor: | CALCIUM ION, amylase | | Authors: | Ohtaki, A, Iguchi, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual conversion of substrate specificities of Thermoactinomyces vulgaris R-47 alpha-amylases TVAI and TVAII by site-directed mutagenesis
CARBOHYDR.RES., 338, 2003
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1IZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase 1 mutant enzyme w398v | | Descriptor: | CALCIUM ION, amylase | | Authors: | Ohtaki, A, Iguchi, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutual conversion of substrate specificities of Thermoactinomyces vulgaris R-47 alpha-amylases TVAI and TVAII by site-directed mutagenesis
CARBOHYDR.RES., 338, 2003
|
|
1WMR
 
 | | Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
1WZM
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1X0C
 
 | | Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isopullulanase | | Authors: | Mizuno, M, Tonozuka, T, Yamamura, A, Miyasaka, Y, Akeboshi, H, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-17 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49
J.Mol.Biol., 376, 2008
|
|
1JI1
 
 | | Crystal Structure Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase 1 | | Descriptor: | ALPHA-AMYLASE I, CALCIUM ION | | Authors: | Kamitori, S, Abe, A, Ohtaki, A, Kaji, A, Tonozuka, T, Sakano, Y. | | Deposit date: | 2001-06-28 | | Release date: | 2002-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures and structural comparison of Thermoactinomyces vulgaris R-47 alpha-amylase 1 (TVAI) at 1.6 A resolution and alpha-amylase 2 (TVAII) at 2.3 A resolution.
J.Mol.Biol., 318, 2002
|
|
