6BMA
 
 | | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
5CD2
 
 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | Descriptor: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-02 | | Release date: | 2015-07-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
6C9Z
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, Glycosyl hydrolase, family 31 | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
5D5H
 
 | | Crystal structure of Mycobacterium tuberculosis Topoisomerase I | | Descriptor: | ACETATE ION, DNA topoisomerase 1, GLYCEROL, ... | | Authors: | Tan, K, Cheng, B, Tse-Dinh, Y.C. | | Deposit date: | 2015-08-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold.
J.Mol.Biol., 428, 2016
|
|
6CA1
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6C9X
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6CA3
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6BB9
 
 | | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-4-deoxychorismate lyase, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-17 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2
To Be Published
|
|
6DGI
 
 | |
6DKH
 
 | | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | L-idonate 5-dehydrogenase (NAD(P)(+)), ZINC ION | | Authors: | Tan, K, Evdokimova, E, McChesney, C, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655
To Be Published
|
|
4EAE
 
 | | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e | | Descriptor: | D-MALATE, Lmo1068 protein, SODIUM ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e
To be Published
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
2NR7
 
 | | Structural Genomics, the crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83 | | Descriptor: | Secretion activator protein, putative | | Authors: | Tan, K, Bigelow, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-01 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The crystal structure of putative secretion activator protein from Porphyromonas gingivalis W83
To be Published
|
|
2O3G
 
 | | Structural Genomics, the crystal structure of a conserved putative domain from Neisseria meningitidis MC58 | | Descriptor: | 1,2-ETHANEDIOL, Putative protein | | Authors: | Tan, K, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of a conserved putative domain from Neisseria meningitidis MC58
To be Published
|
|
2O3F
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
4ESE
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN. | | Descriptor: | DODECAETHYLENE GLYCOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of acyl carrier protein phosphodiesterase from Yersinia pestis CO92 in complex with FMN.
To be Published
|
|
2OF7
 
 | | Structural Genomics, the crystal structure of a tetR-family transcriptional regulator from Streptomyces coelicolor A3 | | Descriptor: | Putative tetR-family transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-02 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of a tetR-family transcriptional regulator from Streptomyces coelicolor A3
To be Published
|
|
2PQQ
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | FORMIC ACID, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2)
To be Published
|
|
2PLS
 
 | | Structural Genomics, the crystal structure of the CorC/HlyC transporter associated domain of a CBS domain protein from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CBS domain protein, ... | | Authors: | Tan, K, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the CorC/HlyC transporter associated domain of a CBS domain protein from Chlorobium tepidum TLS.
To be Published
|
|
2OQT
 
 | | Structural Genomics, the crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS | | Descriptor: | Hypothetical protein SPy0176 | | Authors: | Tan, K, Wu, R, Osipiuk, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of a putative PTS IIA domain from Streptococcus pyogenes M1 GAS
To be Published
|
|
2P0T
 
 | | Structural Genomics, the crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, UPF0307 protein PSPTO_4464 | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000
To be Published
|
|
2P0S
 
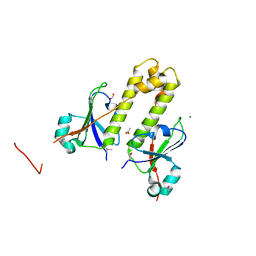 | | Structural Genomics, the crystal structure of a putative ABC transporter domain from Porphyromonas gingivalis W83 | | Descriptor: | ABC transporter, permease protein, putative, ... | | Authors: | Tan, K, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a putative ABC transporter domain from Porphyromonas gingivalis W83
To be Published
|
|
2PMA
 
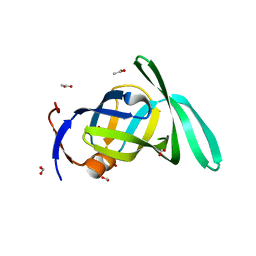 | | Structural Genomics, the crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1. | | Descriptor: | ACETATE ION, FORMIC ACID, Uncharacterized protein | | Authors: | Tan, K, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The crystal structure of a protein Lpg0085 with unknown function (DUF785) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To be Published
|
|
2PKH
 
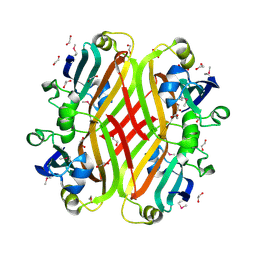 | | Structural Genomics, the crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Histidine utilization repressor | | Authors: | Tan, K, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
4FCA
 
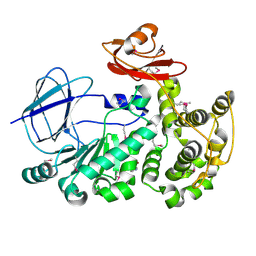 | | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. | | Descriptor: | Conserved domain protein, IMIDAZOLE, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames.
To be Published
|
|
