8SMI
 
 | | Crystal structure of antibody WRAIR-2123 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2123 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SGU
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-13 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
8SMT
 
 | | Crystal structure of antibody WRAIR-2134 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, WRAIR-2134 Fab heavy chain, ... | | Authors: | Sankhala, R.S, Jensen, J.L, Joyce, M.G. | | Deposit date: | 2023-04-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Antibody targeting of conserved sites of vulnerability on the SARS-CoV-2 spike receptor-binding domain.
Structure, 32, 2024
|
|
2C2N
 
 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
2CG5
 
 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase in complex with cytosolic acyl carrier protein and coenzyme A | | Descriptor: | COENZYME A, FATTY ACID SYNTHASE, L-AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, ... | | Authors: | Bunkoczi, G, Joshi, A, Papagrigoriu, E, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Smith, S, Oppermann, U. | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and Substrate Recognition of Human Holo Acp Synthase.
Chem.Biol., 14, 2007
|
|
7KGS
 
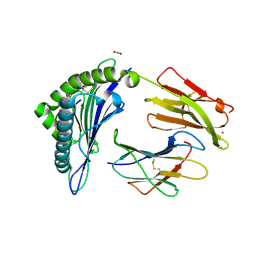 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N138-146 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGT
 
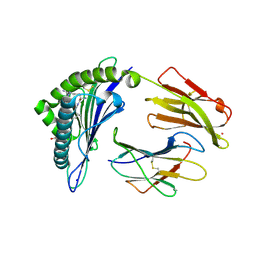 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7KGP
 
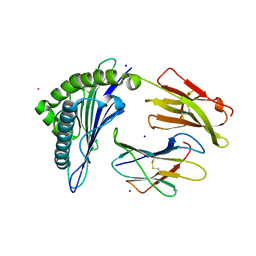 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N316-324 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGO
 
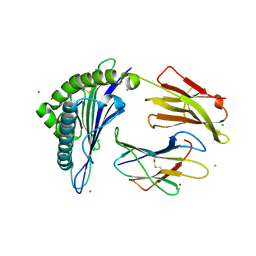 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7LGD
 
 | | HLA-B*07:02 in complex with SARS-CoV-2 nucleocapsid peptide N105-113 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7LGT
 
 | | HLA-B*07:02 in complex with 229E-derived coronavirus nucleocapsid peptide N75-83 | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7KGR
 
 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N159-167 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Nucleoprotein | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KGQ
 
 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N222-230 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7T47
 
 | | KRAS G12D (GppCp) with MRTX-1133 | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Thomas, N.C, Gunn, R.J, Lawson, J.D, Wang, X, Matthew, M.A. | | Deposit date: | 2021-12-09 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A Non-covalent KRASG12D Allele Specific Inhibitor Demonstrates Potent Inhibition of KRAS-dependent Signaling and Regression of KRASG12D-mutant Tumors
Nature, 2022
|
|
4YZU
 
 | |
4Z0K
 
 | |
6C0W
 
 | | Cryo-EM structure of human kinetochore protein CENP-N with the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Centromere protein N, Histone H2A, ... | | Authors: | Zhou, K, Pentakota, S, Vetter, I.R, Morgan, G.P, Petrovic, A, Musacchio, A, Luger, K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Decoding the centromeric nucleosome through CENP-N.
Elife, 6, 2017
|
|
1FGS
 
 | | FOLYLPOLYGLUTAMATE SYNTHETASE FROM LACTOBACILLUS CASEI | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHETASE, MAGNESIUM ION, PYROPHOSPHATE 2- | | Authors: | Sun, X, Bognar, A, Baker, E, Smith, C. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3LEZ
 
 | |
1FC5
 
 | |
3LZ6
 
 | | Guinea Pig 11beta hydroxysteroid dehydrogenase with PF-877423 | | Descriptor: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-adamantan-2-yl-1-ethyl-D-prolinamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pauly, T.A. | | Deposit date: | 2010-03-01 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The development and SAR of pyrrolidine carboxamide 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4ZVJ
 
 | | Structure of human triose phosphate isomerase K13M | | Descriptor: | POTASSIUM ION, SODIUM ION, Triosephosphate isomerase | | Authors: | Amrich, C.G, Smith, C, Heroux, A, VanDemark, A.P. | | Deposit date: | 2015-05-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6996 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim. Biophys. Acta, 1852, 2015
|
|
1KON
 
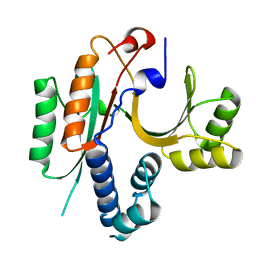 | | CRYSTAL STRUCTURE OF E.COLI YEBC | | Descriptor: | Protein yebC | | Authors: | Jia, J, Smith, C, Lunin, V.V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-12-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | UNPUBLISHED
TO BE PUBLISHED
|
|
6ZH0
 
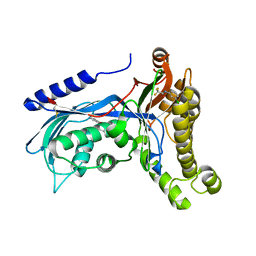 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
1CMV
 
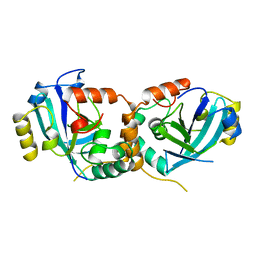 | | HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE | | Authors: | Shieh, H.-S, Kurumbail, R.G, Stevens, A.M, Stegeman, R.A, Sturman, E.J, Pak, J.Y, Wittwer, A.J, Palmier, M.O, Wiegand, R.C, Holwerda, B.C, Stallings, W.C. | | Deposit date: | 1996-08-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Three-dimensional structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
