1VDY
 
 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
2YU4
 
 | | Solution structure of the SP-RING domain in non-SMC element 2 homolog (MMS21, S. cerevisiae) | | Descriptor: | E3 SUMO-protein ligase NSE2, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SP-RING domain in non-SMC element 2 homolog (MMS21, S. cerevisiae)
To be Published
|
|
2YWH
 
 | | Crystal structure of GDP-bound LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein LepA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus.
To be Published
|
|
2Z0V
 
 | | Crystal structure of BAR domain of Endophilin-III | | Descriptor: | SH3-containing GRB2-like protein 3 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of BAR domain of Endophilin-III
To be Published
|
|
2YQL
 
 | | Solution structure of the PHD domain in PHD finger protein 21A | | Descriptor: | PHD finger protein 21A, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain in PHD finger protein 21A
To be Published
|
|
2YT9
 
 | | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278 | | Descriptor: | ZINC ION, zinc finger-containing protein 1 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 345 in Zinc finger protein 278
To be Published
|
|
2YQP
 
 | | Solution structure of the zf-HIT domain in DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 | | Descriptor: | Probable ATP-dependent RNA helicase DDX59, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-HIT domain in DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
To be Published
|
|
2Z0O
 
 | | Crystal structure of APPL1-BAR-PH domain | | Descriptor: | DCC-interacting protein 13-alpha | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of APPL1-BAR-PH domain
To be Published
|
|
2YQR
 
 | | Solution structure of the KH domain in KIAA0907 protein | | Descriptor: | KIAA0907 protein | | Authors: | Kadirvel, S, He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KH domain in KIAA0907 protein
To be Published
|
|
2YT6
 
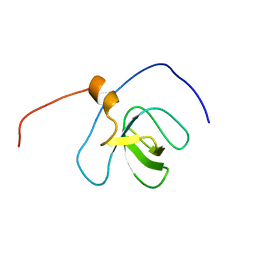 | | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | | Descriptor: | Adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530076O17 product:Yamaguchi sarcoma viral (v-yes) oncogene homolog | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3_1 domain of Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
To be Published
|
|
2YQF
 
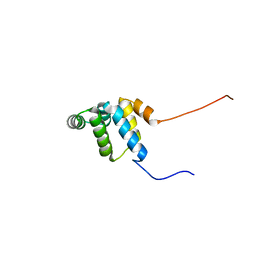 | | Solution structure of the death domain of Ankyrin-1 | | Descriptor: | Ankyrin-1 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the death domain of Ankyrin-1
To be Published
|
|
2YSV
 
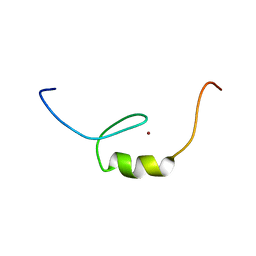 | | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473
To be Published
|
|
2YWQ
 
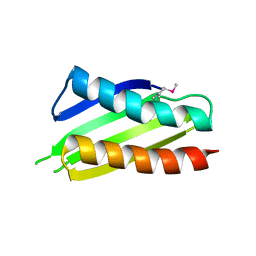 | | Crystal structure of Thermus thermophilus Protein Y N-terminal domain | | Descriptor: | Ribosomal subunit interface protein | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Tatsuguchi, A, Saito, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-21 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of Thermus thermophilus Protein Y N-terminal domain
To be Published
|
|
2Z0K
 
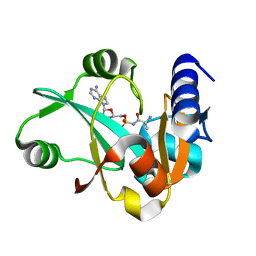 | | Crystal structure of ProX-AlaSA complex from T. thermophilus | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, Putative uncharacterized protein TTHA1699 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ProX-AlaSA complex from T. thermophilus
to be published
|
|
2Z0X
 
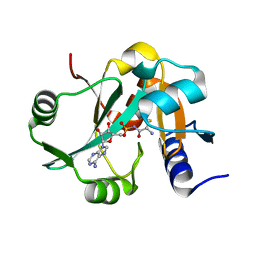 | | Crystal structure of ProX-CysSA complex from T. thermophilus | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Putative uncharacterized protein TTHA1699 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of ProX-CysSA complex from T. thermophilus
To be Published
|
|
2RNE
 
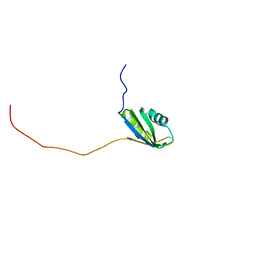 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
2YTA
 
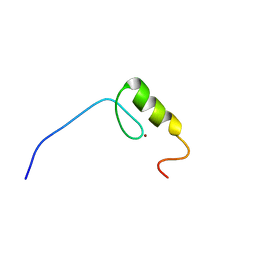 | | Solution structure of C2H2 type Zinc finger domain 3 in Zinc finger protein 32 | | Descriptor: | ZINC ION, Zinc finger protein 32 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 3 in Zinc finger protein 32
To be Published
|
|
2RRA
 
 | | Solution structure of RNA binding domain in human Tra2 beta protein in complex with RNA (GAAGAA) | | Descriptor: | 5'-R(*GP*AP*AP*GP*AP*A)-3', cDNA FLJ40872 fis, clone TUTER2000283, ... | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
2YWK
 
 | | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11 | | Descriptor: | Putative RNA-binding protein 11 | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Uchikubo-Kamo, T, Nishino, A, Morita, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of RRM-domain derived from human putative RNA-binding protein 11
To be Published
|
|
2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
2YQK
 
 | | Solution structure of the SANT domain in Arginine-glutamic acid dipeptide (RE) repeats | | Descriptor: | Arginine-glutamic acid dipeptide repeats protein | | Authors: | Kadirvel, S, He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SANT domain in Arginine-glutamic acid dipeptide (RE) repeats
To be Published
|
|
2YTC
 
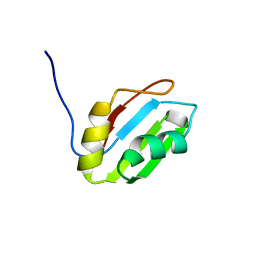 | | Solution structure of RNA binding domain in Pre-mRNA-splicing factor RBM22 | | Descriptor: | Pre-mRNA-splicing factor RBM22 | | Authors: | Kasahara, N, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA binding domain in Pre-mRNA-splicing factor RBM22
To be Published
|
|
2YUD
 
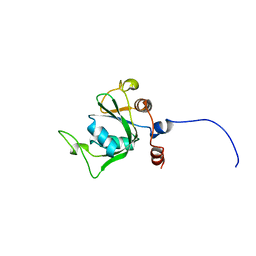 | | Solution structure of the YTH domain in YTH domain-containing protein 1 (Putative splicing factor YT521) | | Descriptor: | YTH domain-containing protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the YTH domain in YTH domain-containing protein 1 (Putative splicing factor YT521)
To be Published
|
|
2YWF
 
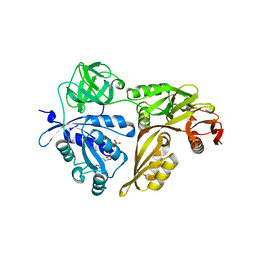 | | Crystal structure of GMPPNP-bound LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein lepA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus
To be Published
|
|
2YQQ
 
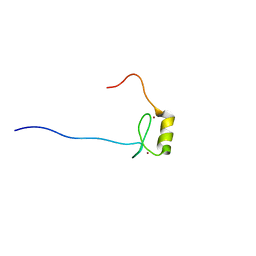 | | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3) | | Descriptor: | ZINC ION, Zinc finger HIT domain-containing protein 3 | | Authors: | Suzuki, S, He, F, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zf-HIT domain in zinc finger HIT domain-containing protein 3 (TRIP-3)
To be Published
|
|
