1DL2
 
 | | CRYSTAL STRUCTURE OF CLASS I ALPHA-1,2-MANNOSIDASE FROM SACCHAROMYCES CEREVISIAE AT 1.54 ANGSTROM RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CLASS I ALPHA-1,2-MANNOSIDASE, ... | | Authors: | Vallee, F, Lipari, F, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 1999-12-08 | | Release date: | 2000-02-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control.
EMBO J., 19, 2000
|
|
6X9M
 
 | |
6X7J
 
 | |
6X7Y
 
 | |
6X8Y
 
 | |
6XAC
 
 | |
6X95
 
 | |
6X7X
 
 | |
6X7T
 
 | |
6XAQ
 
 | |
6X7Z
 
 | | Inositol-bound structure of Marinomonas primoryensis PA14 carbohydrate-binding domain | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, Antifreeze protein, ... | | Authors: | Guo, S, Davies, P.L. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis of Ligand Selectivity by a Bacterial Adhesin Lectin Involved in Multispecies Biofilm Formation.
Mbio, 12, 2021
|
|
6X9P
 
 | |
6X8A
 
 | |
6X8D
 
 | |
6XA5
 
 | |
6WJA
 
 | |
6X5V
 
 | |
6X5W
 
 | |
6X6Q
 
 | |
6X6M
 
 | |
7MSI
 
 | |
6XI3
 
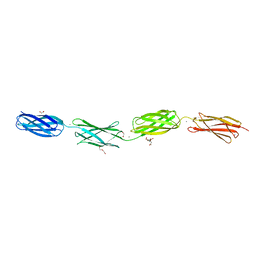 | | Crystal structure of tetra-tandem repeat in extending region of large adhesion protein | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ye, Q, Vance, T.D.R, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
9F2K
 
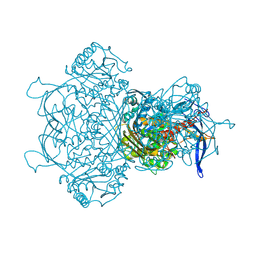 | | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, inositol-3-phosphate synthase | | Authors: | Traeger, T.K, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VW5
 
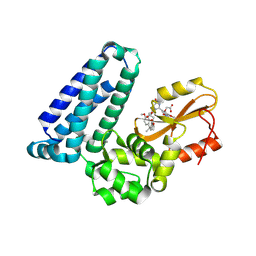 | | Crystal structure of Cbl-b TKB bound to compound 2 | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL-B, MAGNESIUM ION, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
8VW4
 
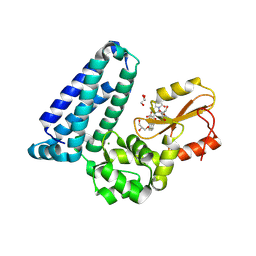 | | Crystal structure of Cbl-b TKB bound to compound 26 | | Descriptor: | (7-methoxy-2-{2-[(1S,3S,4S)-3-(3-methoxy-2-methyl-5-nitrophenyl)-1-methyl-5-oxo-1,5-dihydroimidazo[1,5-a]pyridin-2(3H)-yl]-2-oxoethoxy}quinolin-8-yl)acetic acid, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Yu, C, Murray, J, Hsu, P.L. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Acs Med.Chem.Lett., 15, 2024
|
|
