4LX5
 
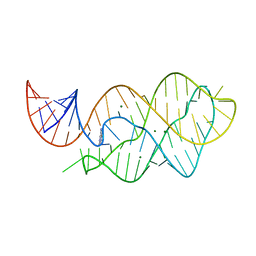 | | X-ray crystal structure of the M6" riboswitch aptamer bound to pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA) | | Descriptor: | MAGNESIUM ION, Mutated adenine riboswitch aptamer, pyrimido[4,5-d]pyrimidine-2,4-diamine | | Authors: | Dunstan, M.S, Leys, D. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species.
J.Am.Chem.Soc., 136, 2014
|
|
2H6C
 
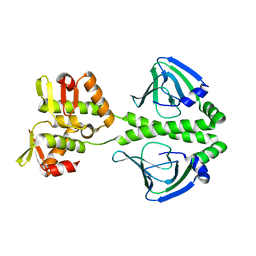 | |
4J34
 
 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
2HJB
 
 | |
2I0S
 
 | |
4L2H
 
 | | Structure of a catalytically inactive PARG in complex with a poly-ADP-ribose fragment | | Descriptor: | Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Brassington, A, Dunstan, M.S, Leys, D. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities.
Nat Commun, 4, 2013
|
|
2HKR
 
 | |
2FW5
 
 | | Diheme cytochrome c from Rhodobacter sphaeroides | | Descriptor: | DHC, diheme cytochrome c, HEME C | | Authors: | Mowat, C.G, Leys, D. | | Deposit date: | 2006-02-01 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies on DHC, the Diheme Cytochrome c from Rhodobacter sphaeroides, and Its Interaction with SHP, the sphaeroides Heme Protein
Biochemistry, 45, 2006
|
|
2IJ4
 
 | | Structure of the A264K mutant of cytochrome P450 BM3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
2IJ5
 
 | |
2IUP
 
 | |
2IJ3
 
 | | Structure of the A264H mutant of cytochrome P450 BM3 | | Descriptor: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
2I0T
 
 | |
2IJ7
 
 | | Structure of Mycobacterium tuberculosis CYP121 in complex with the antifungal drug fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roujeinikova, A, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode.
J.Biol.Chem., 281, 2006
|
|
2IJ2
 
 | | Atomic structure of the heme domain of flavocytochrome P450-BM3 | | Descriptor: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Helen, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
2IUQ
 
 | |
2IUV
 
 | |
2IUR
 
 | |
2H6B
 
 | |
2HJ4
 
 | |
2HKM
 
 | |
2HXC
 
 | |
2I0R
 
 | |
6EVB
 
 | | Structure of E282Q A. niger Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
6EV5
 
 | | Crystal structure of E282Q A. niger Fdc1 with prFMN in the hydroxylated form | | Descriptor: | Ferulic acid decarboxylase 1, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
