5Z5A
 
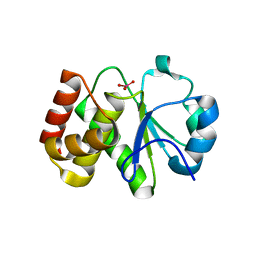 | | Crystal structure of Tk-PTP in the active form | | Descriptor: | Protein-tyrosine phosphatase, VANADATE ION | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
2XHS
 
 | |
9DK8
 
 | | TRIF TIR Filament Cryo-EM Structure | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Manik, M.K, Xiao, L, Wu, H. | | Deposit date: | 2024-09-08 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for TIR domain-mediated innate immune signaling by Toll-like receptor adaptors TRIF and TRAM.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5Z5B
 
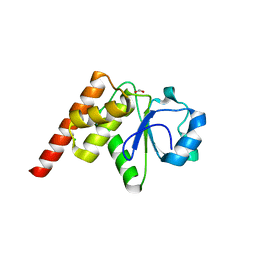 | | Crystal structure of Tk-PTP in the G95A mutant form | | Descriptor: | CHLORIDE ION, FORMIC ACID, Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
1LAM
 
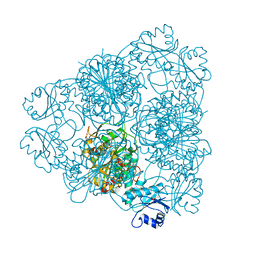 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
8VH5
 
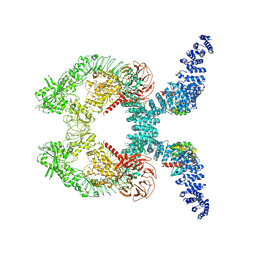 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH4
 
 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
9DLG
 
 | | CryoEM structure of the TIR domain from human TRAM | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Hedger, A, Pospich, S, Pan, M, Gu, W, Ve, T, Raunser, S, Landsberg, M, Nanson, J.D, Kobe, B. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis for TIR domain-mediated innate immune signaling by Toll-like receptor adaptors TRIF and TRAM.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9DYZ
 
 | |
9DZ1
 
 | | VVGGVVGG cyclic peptide | | Descriptor: | VVGGVVGG-cyclic | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2024-10-15 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Formation of rippled beta-sheets from mixed chirality linear and cyclic peptides-new structural motifs based on the pauling-corey rippled beta-sheet.
Chem Sci, 16, 2025
|
|
9DYW
 
 | |
9DYY
 
 | | peptide of mixed chirality, VVGG(DVA)(DVA), assembles into a rippled beta sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, VVGG(DVA)(DVA) | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2024-10-15 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Formation of rippled beta-sheets from mixed chirality linear and cyclic peptides-new structural motifs based on the pauling-corey rippled beta-sheet.
Chem Sci, 16, 2025
|
|
9DYX
 
 | | racemic mixture of peptide VVGGVV forms rippled sheets | | Descriptor: | VVGGVV-amidated racemic mixture, pentafluoropropanoic acid | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2024-10-15 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Formation of rippled beta-sheets from mixed chirality linear and cyclic peptides-new structural motifs based on the pauling-corey rippled beta-sheet.
Chem Sci, 16, 2025
|
|
9DZ0
 
 | |
3OUM
 
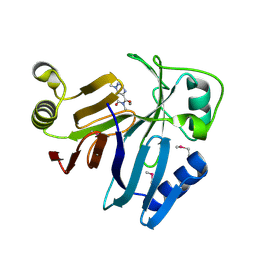 | | Crystal Structure of toxoflavin-degrading enzyme in complex with toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, MANGANESE (II) ION, toxoflavin-degrading enzyme | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2010-09-15 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme
Plos One, 6, 2011
|
|
6XNG
 
 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxyheptadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-07-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
9DKI
 
 | |
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
3OUL
 
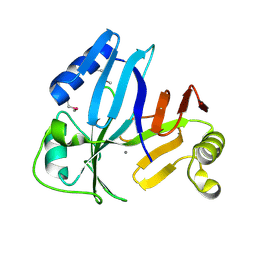 | |
6LBX
 
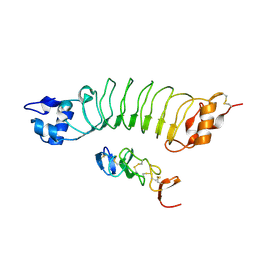 | | Crystal structure of HER2 Domain IV and Rb-H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2, Repebody (Rb-H2) | | Authors: | Cho, H.S, Cha, J.S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Computationally-guided design and affinity improvement of a protein binder targeting a specific site on HER2
Comput Struct Biotechnol J, 19, 2021
|
|
1A77
 
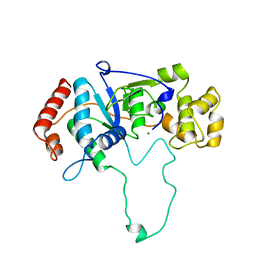 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1A76
 
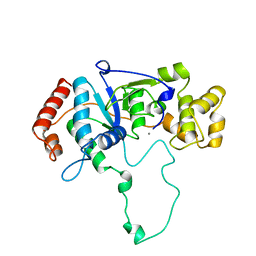 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
6PLN
 
 | |
6USY
 
 | | COAGULATION FACTOR XI CATALYTIC DOMAIN (C123S) IN COMPLEX WITH NVP-XIV936 | | Descriptor: | 1-[(2S)-2-{3-[(3S)-3-amino-2,3-dihydro-1-benzofuran-5-yl]-5-(propan-2-yl)phenyl}-2-hydroxyethyl]-1H-indole-7-carboxylic acid, Coagulation factor XIa light chain | | Authors: | Weihofen, W.A, Clark, K, Nunes, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
