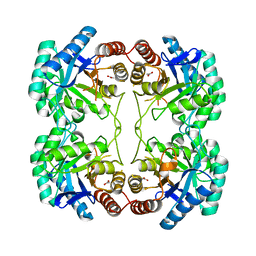
3SZ8
 
 | |
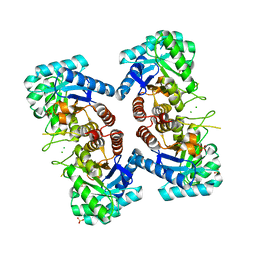
3TML
 
 | |
3TXY
 
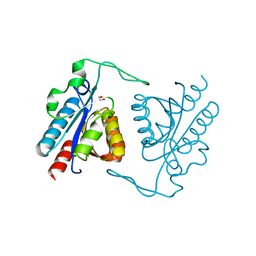 | |
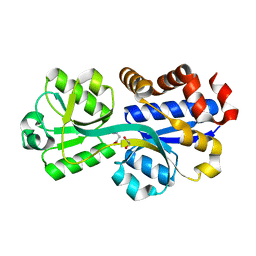
3TMG
 
 | |
3TMQ
 
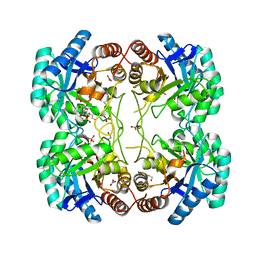 | |
3U7J
 
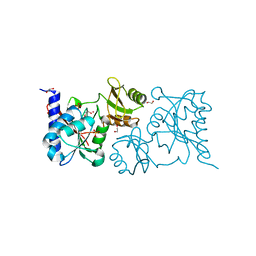 | |
3U0I
 
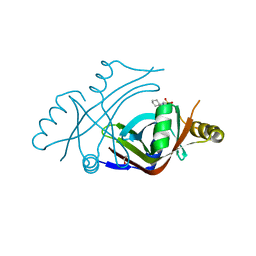 | |
3QHX
 
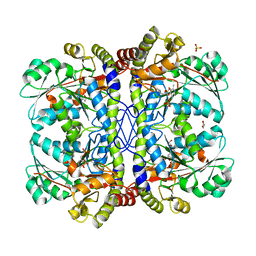 | |
3PXX
 
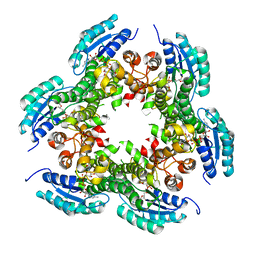 | |
3SX2
 
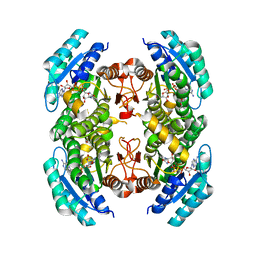 | |
3T7C
 
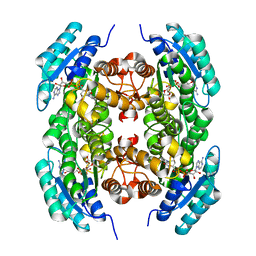 | |
3S55
 
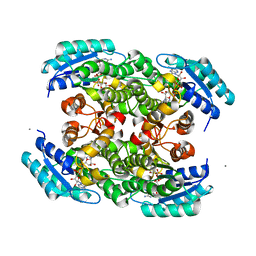 | |
3QH4
 
 | |
3QI6
 
 | |
3U0B
 
 | | Crystal structure of an oxidoreductase from Mycobacterium smegmatis | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family protein, SODIUM ION | | Authors: | Arakaki, T.L, Staker, B.L, Clifton, M.C, Abendroth, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-05 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of FabG4 from Mycolicibacterium smegmatis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3TK1
 
 | |
3TSC
 
 | |
3PXU
 
 | |
3QHD
 
 | |
3Q8H
 
 | | Crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from burkholderia pseudomallei in complex with cytidine derivative EBSI01028 | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-deoxy-5'-[(imidazo[2,1-b][1,3]thiazol-5-ylcarbonyl)amino]cytidine, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytidine derivatives as IspF inhibitors of Burkolderia pseudomallei.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
