6MC1
 
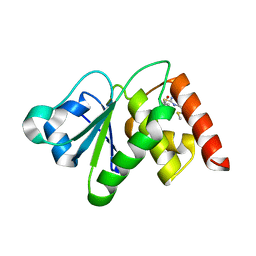 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
9BT8
 
 | | Structure of Src in complex with beta-arrestin 1 revealing SH3 binding sites | | Descriptor: | Antibody fragment Fab30, heavy chain, light chain, ... | | Authors: | Pakharukova, N, Bansia, H, Bassford, D.K, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2024-05-14 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy.
Biorxiv, 2024
|
|
9CX9
 
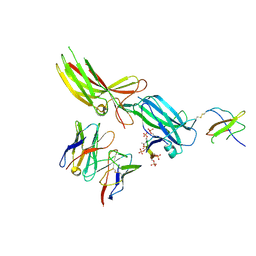 | | Structure of SH3 domain of Src in complex with beta-arrestin 1 | | Descriptor: | Antibody fragment Fab30, heavy chain, light chain, ... | | Authors: | Pakharukova, N, Bansia, H, Lefkowitz, R.J, des Georges, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy.
Biorxiv, 2024
|
|
1ZA7
 
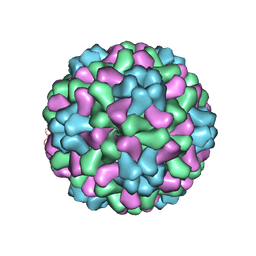 | | The crystal structure of salt stable cowpea cholorotic mottle virus at 2.7 angstroms resolution. | | Descriptor: | Coat protein | | Authors: | Bothner, B, Speir, J.A, Qu, C, Willits, D.A, Young, M.J, Johnson, J.E. | | Deposit date: | 2005-04-05 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics.
J.Virol., 80, 2006
|
|
4RPA
 
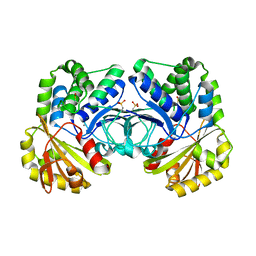 | |
7EBZ
 
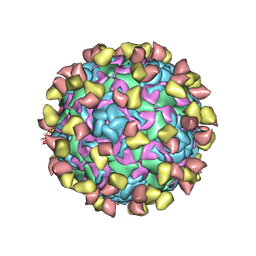 | | EV-D68 in complex with 2H12 Fab (state S1) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7ECY
 
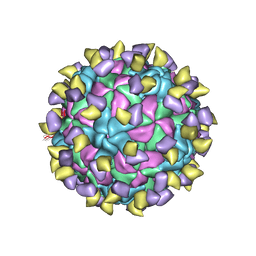 | | EV-D68 in complex with 2H12 Fab (State 3) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EC5
 
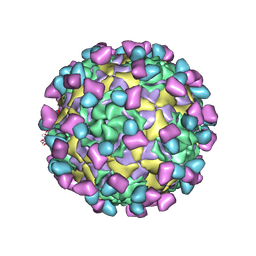 | | EV-D68 in complex with 8F12 Fab | | Descriptor: | 8F12 Fab heavy chain, 8F12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
7EBR
 
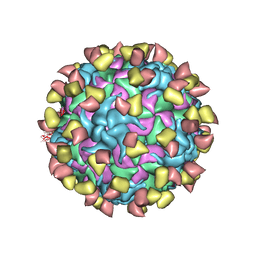 | | EV-D68 in complex with 2H12 Fab (state S2) | | Descriptor: | 2H12 Fab heavy chain, 2H12 Fab light chain, Capsid protein VP1, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural characterization of a two-MAb cocktail for delayed treatment of enterovirus D68 infections.
Nat Commun, 12, 2021
|
|
4FYN
 
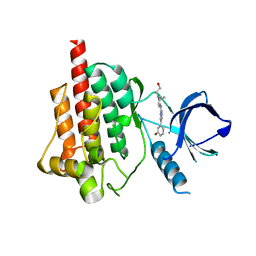 | |
4P83
 
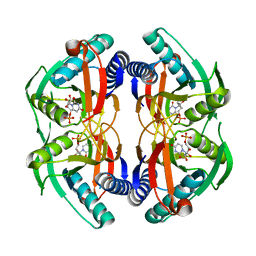 | | Structure of engineered PyrR protein (PURPLE PyrR) | | Descriptor: | Engineered PyrR protein (Purple), URIDINE-5'-MONOPHOSPHATE | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
7EOS
 
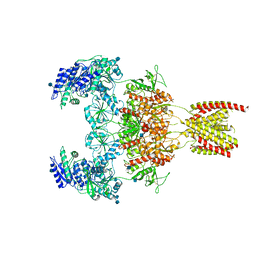 | |
7EOT
 
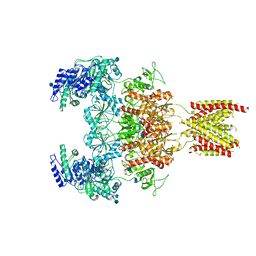 | |
7EOU
 
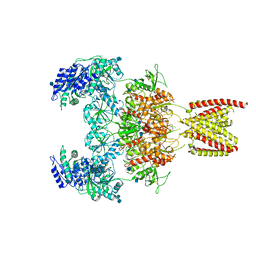 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901/9-AA bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, 9-AMINOACRIDINE, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
7EOQ
 
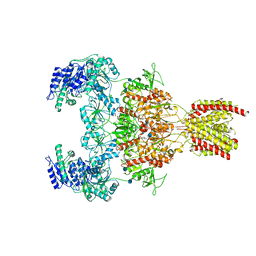 | |
7EOR
 
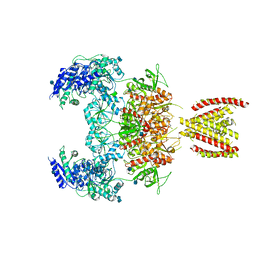 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
4GQ9
 
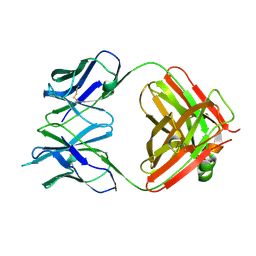 | | Chikungunya virus neutralizing antibody 9.8B Fab fragment | | Descriptor: | Chikungunya virus neutralizing antibody 9.8B Fab fragment heavy chain, Chikungunya virus neutralizing antibody 9.8B Fab fragment light chain | | Authors: | Sun, S, Rossmann, M.G. | | Deposit date: | 2012-08-22 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
4PH2
 
 | | mature N-terminal domain of capsid protein from bovine leukemia virus | | Descriptor: | BLV capsid - N-terminal domain, GLYCEROL, SULFATE ION | | Authors: | Trajtenberg, F, Obal, G, Pritsch, O, Buschiazzo, A. | | Deposit date: | 2014-05-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | STRUCTURAL VIROLOGY. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography.
Science, 349, 2015
|
|
4P81
 
 | | Structure of ancestral PyrR protein (AncORANGEPyrR) | | Descriptor: | Ancestral PyrR protein (Orange), GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P82
 
 | | Structure of PyrR protein from Bacillus subtilis | | Descriptor: | Bifunctional protein PyrR, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
8KIH
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Phomactin Platelet Activating Factor Antagonist Requires a Two-Enzyme Cascade.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8KI5
 
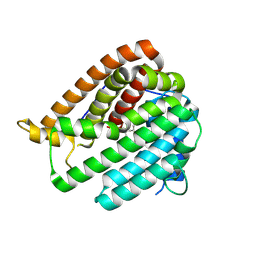 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Phomactin Platelet Activating Factor Antagonist Requires a Two-Enzyme Cascade.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4PH1
 
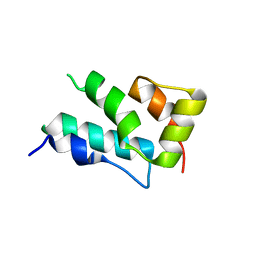 | |
4PH3
 
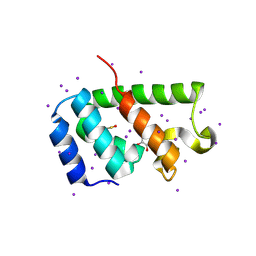 | | N-terminal domain of the capsid protein from bovine leukaemia virus (with no beta-hairpin) | | Descriptor: | BLV capsid, GLYCEROL, IODIDE ION | | Authors: | Trajtenberg, F, Obal, G, Pritsch, O, Buschiazzo, A. | | Deposit date: | 2014-05-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | STRUCTURAL VIROLOGY. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography.
Science, 349, 2015
|
|
7BEG
 
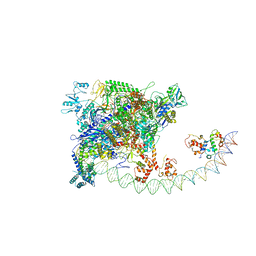 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
