8FIH
 
 | |
5MSQ
 
 | |
8FJE
 
 | | The five-repeat design E8 | | Descriptor: | E8 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJF
 
 | | The three-repeat design H10 | | Descriptor: | H10 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FJG
 
 | | The two-repeat design H12 | | Descriptor: | H12 | | Authors: | Bera, A.K, An, L, Baker, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Hallucination of closed repeat proteins containing central pockets.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8F5I
 
 | | SARS-CoV-2 S2 helix epitope scaffold bound by antibody DH1057.1 | | Descriptor: | DH1057.1 HC, DH1057.1 LC, Specialized acyl carrier protein, ... | | Authors: | Kapingidza, A.B, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-11-14 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
4ZIB
 
 | | Crystal Structure of the C-terminal domain of PylRS mutant bound with 3-benzothienyl-l-alanine and ATP | | Descriptor: | 1,2-ETHANEDIOL, 3-(1-benzothiophen-3-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nakamura, A, Guo, L.T, Wang, Y.S, Soll, D. | | Deposit date: | 2015-04-28 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog.
Nucleic Acids Res., 43, 2015
|
|
8F5H
 
 | |
8FDO
 
 | | SARS-CoV-2 fusion peptide epitope scaffold FP15 bound to DH1058 | | Descriptor: | DH1058 Heavy chain, DH1058 Light chain, FP15 | | Authors: | Kapingidza, A.B, Marston, D.J, Wrapp, D, Winters, K, Azoitei, M.L. | | Deposit date: | 2022-12-04 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered immunogens to elicit antibodies against conserved coronavirus epitopes.
Nat Commun, 14, 2023
|
|
3MES
 
 | | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, DECAMETHONIUM ION, ... | | Authors: | Qiu, W, Wernimont, A, Hills, T, Lew, J, Artz, J.D, Xiao, T, Allali-Hassani, A, Vedadi, M, Kozieradzki, I, Cossar, D, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Hui, R, Ma, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030
TO BE PUBLISHED
|
|
4XCG
 
 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Smits, C, Chaston, J.J, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.737 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
4ZA7
 
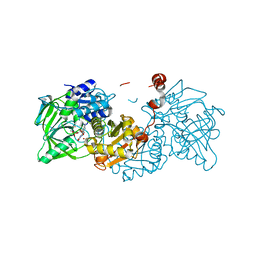 | | Structure of A. niger Fdc1 in complex with alpha-methyl cinnamic acid | | Descriptor: | (2E)-2-methyl-3-phenylprop-2-enoic acid, 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAN
 
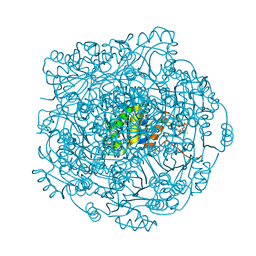 | |
5MSD
 
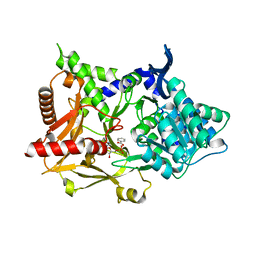 | |
4ZAX
 
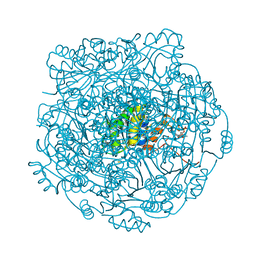 | | Structure of UbiX in complex with oxidised prenylated FMN (radical) | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
5MQX
 
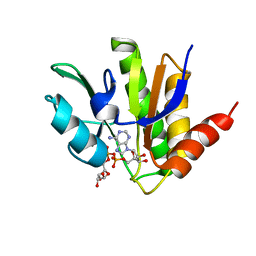 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
1ASY
 
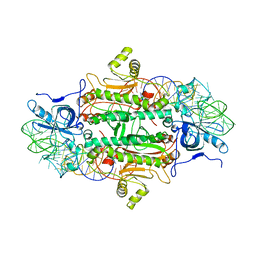 | | CLASS II AMINOACYL TRANSFER RNA SYNTHETASES: CRYSTAL STRUCTURE OF YEAST ASPARTYL-TRNA SYNTHETASE COMPLEXED WITH TRNA ASP | | Descriptor: | ASPARTYL-tRNA SYNTHETASE, T-RNA (75-MER) | | Authors: | Ruff, M, Cavarelli, J, Rees, B, Krishnaswamy, S, Thierry, J.C, Moras, D. | | Deposit date: | 1995-01-19 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp).
Science, 252, 1991
|
|
4ZOT
 
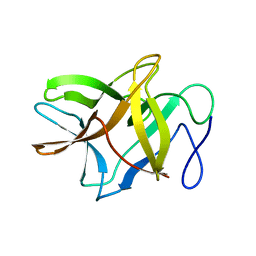 | | Crystal structure of BbKI, a disulfide-free plasma kallikrein inhibitor at 1.4 A resolution | | Descriptor: | Kunitz-type serine protease inhibitor BbKI | | Authors: | Shabalin, I.G, Zhou, D, Wlodawer, A, Oliva, M.L.V. | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of BbKI, a disulfide-free plasma kallikrein inhibitor.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1GJY
 
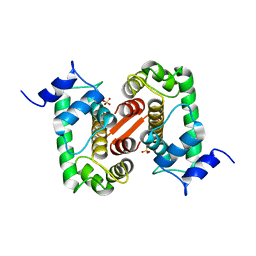 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
4ZA0
 
 | | Structure of Human Enolase 2 in complex with Phosphonoacetohydroxamate | | Descriptor: | Gamma-enolase, MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
4ZAD
 
 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4ZAV
 
 | | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
6RNJ
 
 | | TR-SMX closed state structure (0-5ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, F, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
3AGL
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1039 | | Descriptor: | (10R,20R,23R)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamido propyl)-10-methyl-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Ragozina, J, Uri, A, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-04-02 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
4ZAA
 
 | | Structure of A. niger Fdc1 in complex with 4-vinyl guaiacol | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, 2-Methoxy-4-vinylphenol, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
