4XJZ
 
 | |
4XOG
 
 | |
9D11
 
 | | Smarca2 Bromodomain in complex with compound 22 | | Descriptor: | (12'S)-4'-chloro-10'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Discovery of High-Affinity SMARCA2/4 Bromodomain Ligands and Development of Potent and Exceptionally Selective SMARCA2 PROTAC Degraders.
J.Med.Chem., 68, 2025
|
|
9D12
 
 | | Smarca2 Bromodomain in complex with compound 15 | | Descriptor: | (12'R)-4'-chloro-9'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Discovery of High-Affinity SMARCA2/4 Bromodomain Ligands and Development of Potent and Exceptionally Selective SMARCA2 PROTAC Degraders.
J.Med.Chem., 68, 2025
|
|
4XIO
 
 | |
4XJU
 
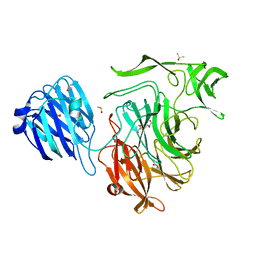 | | Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 4-acetamido-2-fluoro-3-hydroxy-6-[1,2-dihydroxyethyl]-7,8-dioxabicyclo[3.2.1]octane-1-carboxylic acid | | Descriptor: | (1R,2R,3R,4R,5R,7R)-2-(acetylamino)-7-[(1R)-1,2-dihydroxyethyl]-4-fluoro-3-hydroxy-6,8-dioxabicyclo[3.2.1]octane-5-carboxylic acid, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Brear, P. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | `The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To be published
|
|
4XHX
 
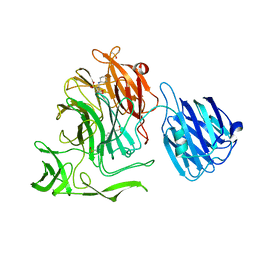 | |
4XIK
 
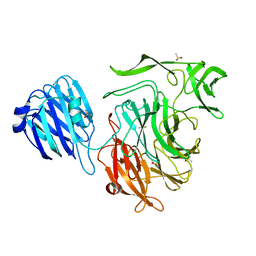 | |
4XJA
 
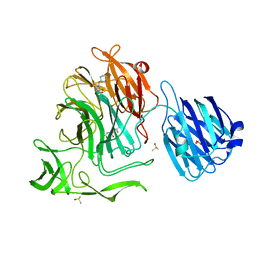 | | Crystal structure of the NanB sialidase from streptococcus pneumoniae in complex with 5-acetamido-2,3-difluoro-3-hydroxy-6-[1,2,3-trihydroxypropyl]oxane-2-carboxylic acid | | Descriptor: | (1s,3R,4S)-1-[(cyclohexylamino)methyl]-3,4-dihydroxycyclopentanesulfonic acid, (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Brear, P. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | `The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To be published
|
|
4XIL
 
 | |
4XMA
 
 | |
4XMI
 
 | |
6JN2
 
 | | Crystal structure of the coiled-coil domains of human DOT1L in complex with AF10 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | Authors: | Song, X, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2019-03-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A higher-order configuration of the heterodimeric DOT1L-AF10 coiled-coil domains potentiates their leukemogenenic activity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P62
 
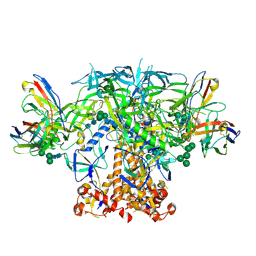 | | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env BG505 NFL TD+, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6PEH
 
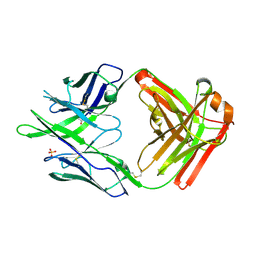 | | Crystal structure of rabbit monoclonal anti-HIV antibody 1C2 | | Descriptor: | 1C2 Fab Heavy Chain, 1C2 Fab Light Chain, SULFATE ION | | Authors: | Liban, T, Pancera, M. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6PKA
 
 | |
6P65
 
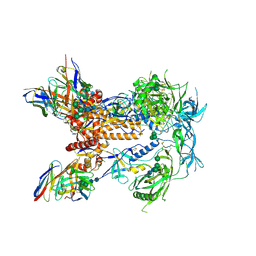 | | HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
6PMD
 
 | |
9E1K
 
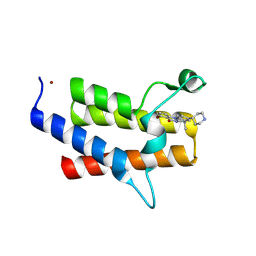 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 11 | | Descriptor: | (12S)-4-bromo-7,7-dimethyl-9-(piperidin-4-yl)indolo[1,2-a]quinazolin-5(7H)-one, Isoform Short of Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E31
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 6 | | Descriptor: | (12'R)-4'-chloro-9'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E30
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 13 | | Descriptor: | (12'S)-4'-chloro-10'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
6OX7
 
 | | The complex of 1918 NS1-ED and the iSH2 domain of the human p85beta subunit of PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Shen, Q, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2019-05-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
9JDC
 
 | | Structure of chanoclavine synthase from Claviceps fusiformis in complex with prechanoclavine | | Descriptor: | (2~{S})-2-(methylamino)-3-[4-[(1~{E})-3-methylbuta-1,3-dienyl]-1~{H}-indol-3-yl]propanoic acid, Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 640, 2025
|
|
9JDB
 
 | | Structure of chanoclavine synthase from Claviceps fusiformis | | Descriptor: | Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 640, 2025
|
|
