3SAK
 
 | |
5X9P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
8ZMF
 
 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
8ZMG
 
 | | Crystal structure of an inverse agonist antipsychotic drug pimavanserin-bound 5-HT2A | | Descriptor: | 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, Pimavanserin | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
5X9O
 
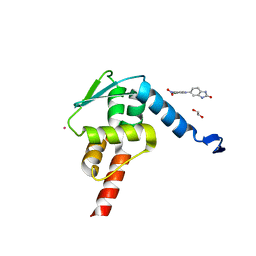 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
1C4E
 
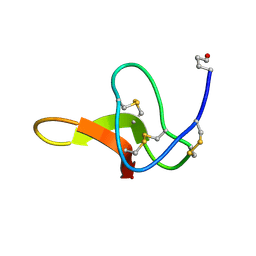 | |
1SAL
 
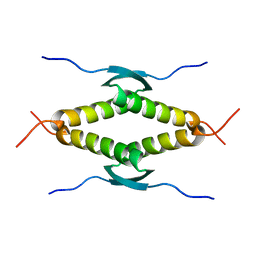 | |
1SAK
 
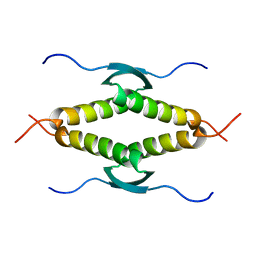 | |
1SAE
 
 | |
1SAF
 
 | |
6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
5XCO
 
 | |
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
7INS
 
 | | STRUCTURE OF PORCINE INSULIN COCRYSTALLIZED WITH CLUPEINE Z | | Descriptor: | GENERAL PROTAMINE CHAIN, INSULIN (CHAIN A), INSULIN (CHAIN B), ... | | Authors: | Balschmidt, P, Hansen, F.B, Dodson, E, Dodson, G, Korber, F. | | Deposit date: | 1991-09-03 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of porcine insulin cocrystallized with clupeine Z.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
5X54
 
 | | Crystal structure of the Keap1 Kelch domain in complex with a tetrapeptide | | Descriptor: | ACE-GLU-TRP-TRP-TRP, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Kelch-like ECH-associated protein 1-inhibitory tetrapeptide and its structural characterization
Biochem. Biophys. Res. Commun., 486, 2017
|
|
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
8YSX
 
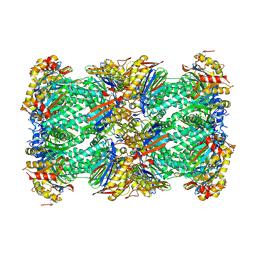 | | canine immunoproteasome 20S subunit in complex with compound 2 | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit beta, Proteasome subunit beta type-8, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
4QCD
 
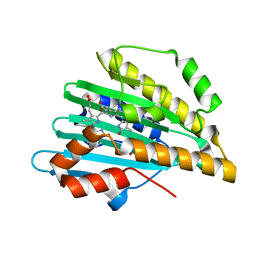 | | Neutron crystal structure of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin IXalpha at room temperature. | | Descriptor: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase, trideuteriooxidanium | | Authors: | Unno, M, Ishikawa-Suto, K, Ishihara, M, Hagiwara, Y, Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.932 Å), X-RAY DIFFRACTION | | Cite: | Insights into the Proton Transfer Mechanism of a Bilin Reductase PcyA Following Neutron Crystallography.
J. Am. Chem. Soc., 137, 2015
|
|
7YK9
 
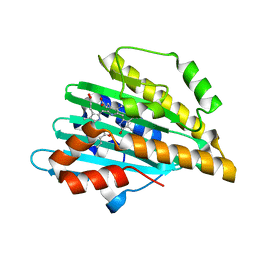 | | Neutron Structure of PcyA I86D Mutant Complexed with Biliverdin at Room Temperature | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Unno, M, Igarashi, K. | | Deposit date: | 2022-07-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography and quantum chemical analysis of bilin reductase PcyA mutants reveal substrate and catalytic residue protonation states.
J.Biol.Chem., 299, 2022
|
|
