4DUH
 
 | | Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor | | Descriptor: | 4-{[4'-methyl-2'-(propanoylamino)-4,5'-bi-1,3-thiazol-2-yl]amino}benzoic acid, DNA gyrase subunit B | | Authors: | Brvar, M, Renko, M, Perdih, A, Solmajer, T, Turk, D. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors.
J.Med.Chem., 55, 2012
|
|
2XE0
 
 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | 24MER DNA, ACETATE ION, I-CREI V2V3 VARIANT, ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Redondo, P, Villate, M, Merino, N, Marenchino, M, D'Abramo, M, Gervasio, F.L, Grizot, S, Daboussi, F, Smith, J, Chion-Sotine, I, Paques, F, Duchateau, P, Alibes, A, Stricher, F, Serrano, L, Blanco, F.J, Montoya, G. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis of Engineered Meganuclease Targeting of the Endogenous Human Rag1 Locus
Nucleic Acids Res., 39, 2011
|
|
2WLQ
 
 | | Nucleophile-disabled Lam16A mutant holds laminariheptaose (L7) in a cyclical conformation | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H, Hansson, H, Engstrom, A, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
4PI9
 
 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4PI7
 
 | | Crystal structure of S. Aureus Autolysin E in complex with disaccharide NAM-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PIA
 
 | | Crystal structure of S. Aureus Autolysin E | | Descriptor: | Autolysin E, CHLORIDE ION | | Authors: | Mihelic, M, Renko, M, Dobersek, A, Bedrac, L, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
2WNE
 
 | | Mutant Laminarinase 16A cyclizes laminariheptaose | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Andersson, E, Widmalm, G, Jonsson, K.H.M, Hansson, H, Engstrom, A, Einarsson, E, Forsberg, Z, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2009-07-09 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Synthesis of Cyclic Beta-Glucan Using Laminarinase 16A Glycosynthase Mutant from the Basidiomycete Phanerochaete Chrysosporium.
J.Am.Chem.Soc., 132, 2010
|
|
4I8D
 
 | | Crystal Structure of Beta-D-glucoside glucohydrolase from Trichoderma reesei | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucoside glucohydrolase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Helmich, K.E, Banerjee, G, Bianchetti, C.M, Gudmundsson, M, Sandgren, M, Walton, J.D, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 beta-Glucosidase, Cel3A from Hypocrea jecorina.
J.Biol.Chem., 289, 2014
|
|
2XSP
 
 | | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, ... | | Authors: | Haddad-momeni, M, Hansson, H, Mikkelsen, N.E, Wang, X, Svedberg, J, Sandgren, M, Stahlberg, J. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of the Glycoside Hydrolase Family 7 Cellobiohydrolase of the Tree-Killing Fungus Heterobasidion Irregulare.
J.Biol.Chem., 288, 2013
|
|
5A7D
 
 | | Tetrameric assembly of LGN with Inscuteable | | Descriptor: | INSCUTEABLE, PINS | | Authors: | Culurgioni, S, Mari, S, Bonetto, G, Gallini, S, Brennich, M, Round, A, Mapelli, M. | | Deposit date: | 2015-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of the Insc:Lgn Tetramer Reveals a New Function of Lgn in Promoting Asymmetric Cell Divisions
To be Published
|
|
5BRV
 
 | | Catalytic Improvement of an Artificial Metalloenzyme by Computational Design | | Descriptor: | Carbonic anhydrase 2, ZINC ION, pentamethylcyclopentadienyl iridium [N-benzensulfonamide-(2-pyridylmethyl-4-benzensulfonamide)amin] chloride | | Authors: | Heinisch, T, Pellizzoni, M, Duerrenberger, M, Tinberg, C.E, Koehler, V, Klehr, J, Haeussinger, D, Baker, D, Ward, T.R. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving the Catalytic Performance of an Artificial Metalloenzyme by Computational Design.
J.Am.Chem.Soc., 137, 2015
|
|
4ALE
 
 | | Structure changes of Polysaccharide monooxygenase CBM33A from Enterococcus faecalis by X-ray induced photoreduction. | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALQ
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
5MDT
 
 | |
5AKF
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE CODING STRAND A AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
4ALS
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALR
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALT
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
5AKN
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE non-CODING STRAND B AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP *CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
2W52
 
 | | 2 beta-glucans (6-O-glucosyl-laminaritriose) in both donor and acceptor sites of GH16 Laminarinase 16A from Phanerochaete chrysosporium. | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Vasur, J, Kawai, R, Andersson, E, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-Ray Crystal Structures of Phanerochaete Chrysosporium Laminarinase 16A in Complex with Products from Lichenin and Laminarin Hydrolysis
FEBS J., 276, 2009
|
|
2W39
 
 | | Glc(beta-1-3)Glc disaccharide in -1 and -2 sites of Laminarinase 16A from Phanerochaete chrysosporium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, PUTATIVE LAMINARINASE, ... | | Authors: | Vasur, J, Kawai, R, Andersson, E, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2008-11-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-Ray Crystal Structures of Phanerochaete Chrysosporium Laminarinase 16A in Complex with Products from Lichenin and Laminarin Hydrolysis
FEBS J., 276, 2009
|
|
5AK9
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3, ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5AKM
 
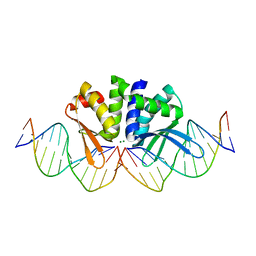 | | THE CRYSTAL STRUCTURE OF I-DMOI G20S IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MG | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5NBS
 
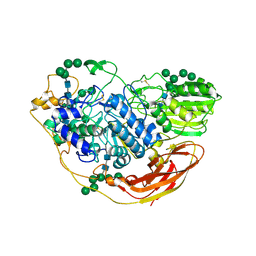 | | Structural studies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Gudmundsson, M, Karkehabadi, S, Kaper, T, Sandgren, M. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of a glycoside hydrolase family 3 beta-glucosidase from the model fungus Neurospora crassa.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
