1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | Authors: | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | Deposit date: | 2004-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
1IWP
 
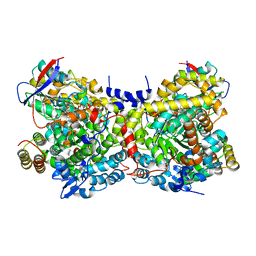 | | Glycerol Dehydratase-cyanocobalamin Complex of Klebsiella pneumoniae | | Descriptor: | COBALAMIN, Glycerol Dehydratase Alpha subunit, Glycerol Dehydratase Beta subunit, ... | | Authors: | Yamanishi, M, Yunoki, M, Tobimatsu, T, Toraya, T. | | Deposit date: | 2002-05-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol.
Eur.J.Biochem., 269, 2002
|
|
1J1J
 
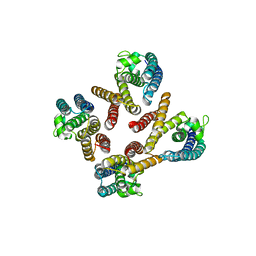 | | Crystal Structure of human Translin | | Descriptor: | Translin | | Authors: | Sugiura, I, Sasaki, C, Hasegawa, T, Kohno, T, Sugio, S, Moriyama, H, Kasai, M, Matsuzaki, T. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human translin at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MKC
 
 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
2DFV
 
 | | Hyperthermophilic threonine dehydrogenase from Pyrococcus horikoshii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable L-threonine 3-dehydrogenase, SULFATE ION, ... | | Authors: | Ishikawa, K, Higashi, N, Nakamura, T, Matsuura, T, Nakagawa, A. | | Deposit date: | 2006-03-03 | | Release date: | 2007-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The first crystal structure of L-threonine dehydrogenase.
J.Mol.Biol., 366, 2007
|
|
2CZR
 
 | | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA | | Descriptor: | GLYCEROL, TBP-interacting protein, ZINC ION | | Authors: | Yamamoto, T, Matsuda, T, Inoue, T, Matsumura, H, Morikawa, M, Kanaya, S, Kai, Y. | | Deposit date: | 2005-07-15 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA
Protein Sci., 15, 2006
|
|
1J1T
 
 | | Alginate lyase from Alteromonas sp.272 | | Descriptor: | Alginate Lyase, CALCIUM ION, SULFATE ION | | Authors: | Motoshima, H, Iwatomo, Y, Watanabe, K, Oda, T, Muramatsu, T. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Alginate Lyase from Alteromonas sp.272
To be published
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
3AYT
 
 | | TTHB071 protein from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3AYV
 
 | | TTHB071 protein from Thermus thermophilus HB8 soaking with ZnCl2 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
1KM7
 
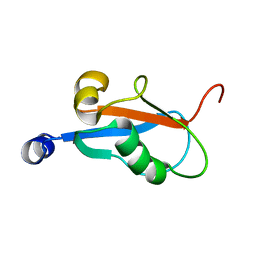 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor Associated Protein | | Descriptor: | GABA(A) Receptor Associated Protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-14 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
3A34
 
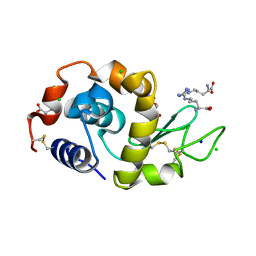 | | Effect of Ariginine on lysozyme | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Matsuura, T, Yamaguchi, H. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Implication of Arg bindings on aromatic surfaces of lysozyme for additives that prevent protein aggregation
TO BE PUBLISHED
|
|
3X2S
 
 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
2D4D
 
 | | The Crystal Structure of human beta2-microglobulin, L39W W60F W95F Mutant | | Descriptor: | Beta-2-microglobulin, SODIUM ION | | Authors: | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | Deposit date: | 2005-10-17 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
2D4F
 
 | | The Crystal Structure of human beta2-microglobulin | | Descriptor: | Beta-2-microglobulin, SODIUM ION | | Authors: | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | Deposit date: | 2005-10-18 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
3VUO
 
 | | Crystal structure of nontoxic nonhemagglutinin subcomponent (NTNHA) from clostridium botulinum serotype D strain 4947 | | Descriptor: | NTNHA | | Authors: | Sagane, Y, Miyashita, S.-I, Miyata, K, Matsumoto, T, Inui, K, Hayashi, S, Suzuki, T, Hasegawa, K, Yajima, S, Yamano, A, Niwa, K, Watanabe, T. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin
Biochem.Biophys.Res.Commun., 425, 2012
|
|
2E3T
 
 | | Crystal structure of rat xanthine oxidoreductase mutant (W335A and F336L) | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Asai, R, Nishino, T, Matsumura, T, Okamoto, K, Pai, E.F, Nishino, T. | | Deposit date: | 2006-11-28 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two mutations convert mammalian xanthine oxidoreductase to highly superoxide-productive xanthine oxidase
J.Biochem.(Tokyo), 141, 2007
|
|
2E0A
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shiromizu, I, Kawamoto, M, Matsusue, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2D6C
 
 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
2DE4
 
 | | Crystal structure of DSZB C27S mutant in complex with biphenyl-2-sulfinic acid | | Descriptor: | 1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE2
 
 | | Crystal structure of desulfurization enzyme DSZB | | Descriptor: | ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, GLYCEROL | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2DE3
 
 | | Crystal structure of DSZB C27S mutant in complex with 2'-hydroxybiphenyl-2-sulfinic acid | | Descriptor: | 2'-HYDROXY-1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B, ... | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
2EKT
 
 | | Crystal structure of myoglobin reconstituted with 6-methyl-6-depropionatehemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
2EKU
 
 | | Crystal structure of myoglobin reconstituted with 7-methyl-7-depropionatehemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
