6EMF
 
 | |
6EN7
 
 | |
6EMG
 
 | |
6XDG
 
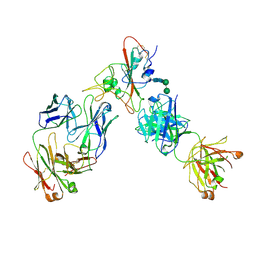 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
6R2M
 
 | | Crystal structure of PssZ from Listeria monocytogenes | | Descriptor: | Glycoside transferase | | Authors: | Wu, H, Cheng, J, Qiao, S, Li, D, Ma, L. | | Deposit date: | 2019-03-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal structure of the glycoside hydrolase PssZ from Listeria monocytogenes.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8XQT
 
 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQR
 
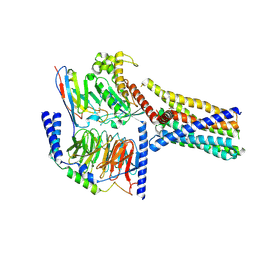 | | Structure 2 of human class T GPCR TAS2R14-miniGs/gust complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQO
 
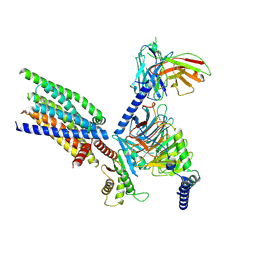 | | Structure of human class T GPCR TAS2R14-Gi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8XQS
 
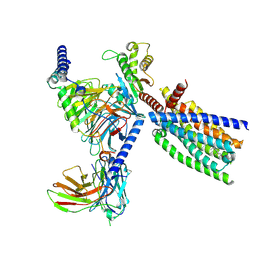 | | Structure of human class T GPCR TAS2R14-DNGi complex with Flufenamic acid. | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8G59
 
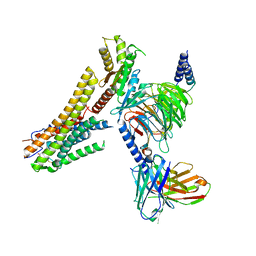 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7AU7
 
 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YLH
 
 | |
6YLE
 
 | |
6YLF
 
 | |
6Y69
 
 | |
6YLY
 
 | | pre-60S State NE2 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YLX
 
 | | pre-60S State NE1 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YLG
 
 | |
2PQ4
 
 | |
2KQX
 
 | |
1F2A
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (II) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONYLMETHYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2B
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (III) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID 4-NITRO-PHENYL ESTER, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
