5O7I
 
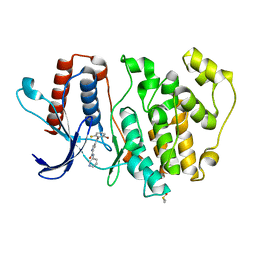 | | ERK5 in complex with a pyrrole inhibitor | | Descriptor: | 4-(2-bromanyl-6-fluoranyl-phenyl)carbonyl-~{N}-pyridin-3-yl-1~{H}-pyrrole-2-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J.A, Heptinstall, A, Myers, S. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
9EAJ
 
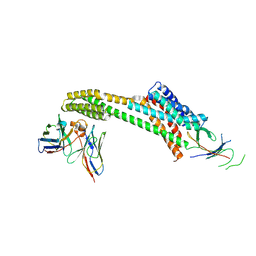 | |
9EAI
 
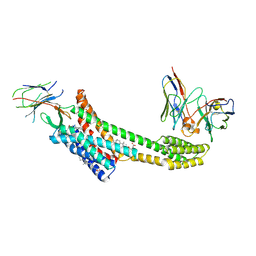 | | Structure of nanobody AT206 in complex with the losartan-bound angiotensin II type I receptor (AT1R) | | Descriptor: | BAG2 Anti-BRIL Fab Heavy Chain, BAG2 Anti-BRIL Fab Light Chain, CHOLESTEROL, ... | | Authors: | Skiba, M.A, Liu, J, Kruse, A.C. | | Deposit date: | 2024-11-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Epitope-directed selection of GPCR nanobody ligands with evolvable function.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9EAH
 
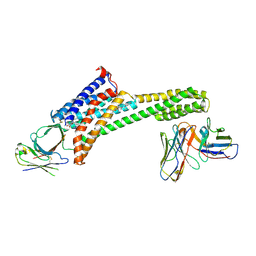 | |
5LRQ
 
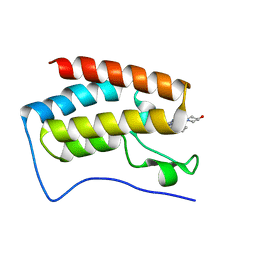 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
1G9O
 
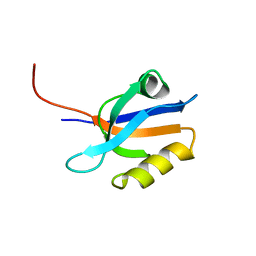 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
3P23
 
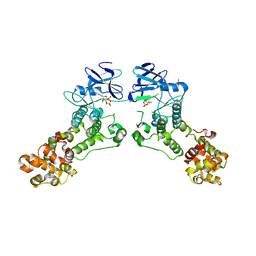 | |
2N3Y
 
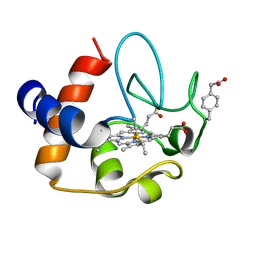 | | NMR structure of the Y48pCMF variant of human cytochrome c in its reduced state | | Descriptor: | Cytochrome c, Mesoheme | | Authors: | Moreno-Beltran, B, Del Conte, R, Diaz-Quintana, A, De la Rosa, M.A, Turano, P, Diaz-Moreno, I. | | Deposit date: | 2015-06-15 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7GV5
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 17.50 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVC
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 3 at 8.70 MGy X-ray dose | | Descriptor: | 5-[(5,6-dichloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GV2
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 13.75 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GUW
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 6.25 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GV0
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 11.25 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVP
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 4 at 5.60 MGy X-ray dose | | Descriptor: | 5-[(2,5-dichloropyridin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GW5
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 5 at 6.45 MGy X-ray dose. | | Descriptor: | 5-[(2-chloro-5-fluoropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GWL
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 6 at 7.38 MGy X-ray dose. | | Descriptor: | 5-{[5-chloro-2-(dimethylamino)pyrimidin-4-yl]amino}-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GUU
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 3.75 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GV8
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 3 at 2.90 MGy X-ray dose | | Descriptor: | 5-[(5,6-dichloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVR
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 4 at 8.40 MGy X-ray dose | | Descriptor: | 5-[(2,5-dichloropyridin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GWO
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 6 at 11.07 MGy X-ray dose. | | Descriptor: | 5-{[5-chloro-2-(dimethylamino)pyrimidin-4-yl]amino}-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GX0
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 7 at 8.70 MGy X-ray dose. | | Descriptor: | 4-chloro-6-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]pyrimidine-5-carbonitrile, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GV4
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 2 at 16.25 MGy X-ray dose. | | Descriptor: | 5-[(5-bromo-2-chloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GWB
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 5 at 14.19 MGy X-ray dose. | | Descriptor: | 5-[(2-chloro-5-fluoropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVH
 
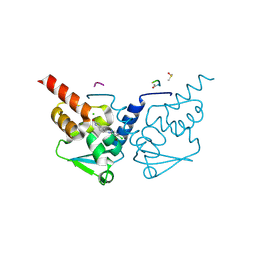 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 3 at 15.95 MGy X-ray dose | | Descriptor: | 5-[(5,6-dichloropyrimidin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GVV
 
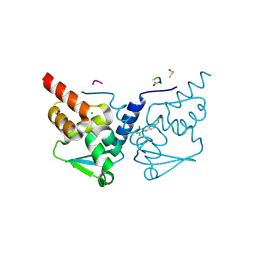 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 4 at 14.00 MGy X-ray dose | | Descriptor: | 5-[(2,5-dichloropyridin-4-yl)amino]-1,3-dihydro-2H-indol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
