6OZ7
 
 | | Putative oxidoreductase from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-15 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | Descriptor: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-06 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
6V3Q
 
 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | Descriptor: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
6U13
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed antibiotic moxalactam.
To Be Published
|
|
6U2Y
 
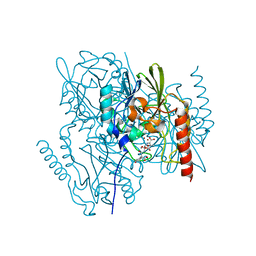 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, 1,2-ETHANEDIOL, NICKEL (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two Ni ions
To Be Published
|
|
6V5M
 
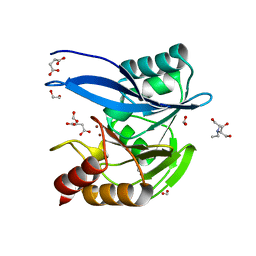 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
6V71
 
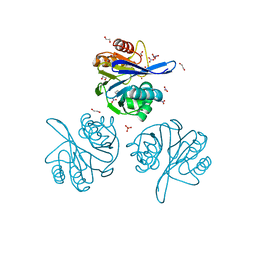 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
6VBB
 
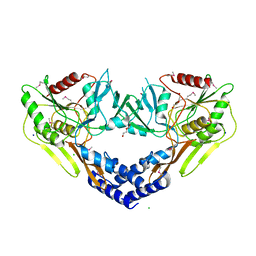 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6UXT
 
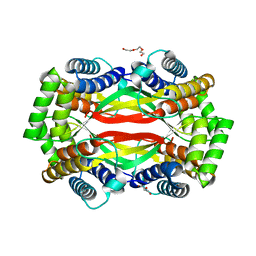 | | Crystal structure of unknown function protein yfdX from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Chang, C, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Crystal structure of unknown function protein yfdX from Shigella flexneri
To Be Published
|
|
6V3U
 
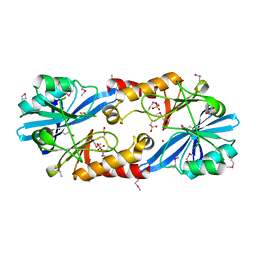 | | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the NDM_FIM-1 like Metallo-beta-Lactamase from Erythrobacter litoralis in the Mono-Zinc Form
To Be Published
|
|
6VBF
 
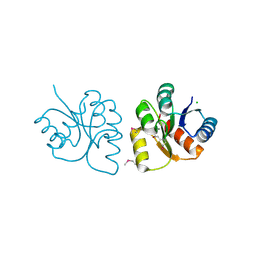 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-18 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
5KP2
 
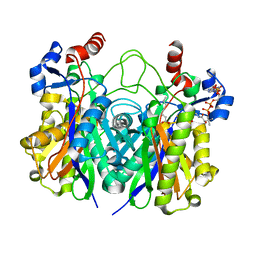 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
5KZ6
 
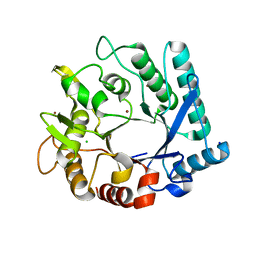 | | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Chitinase, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.
To Be Published
|
|
7TRW
 
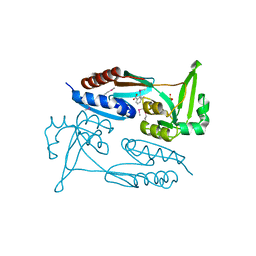 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | Authors: | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TRV
 
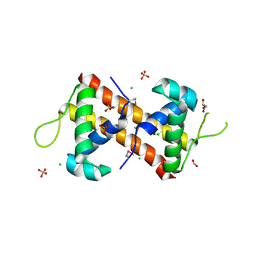 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7TVX
 
 | |
5EAV
 
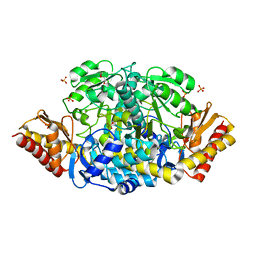 | | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii
To Be Published
|
|
5DZS
 
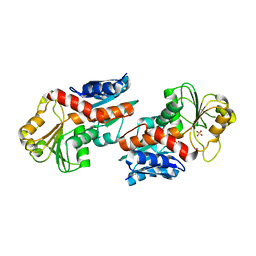 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
5DVY
 
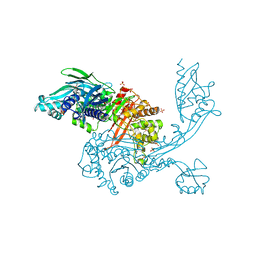 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5E31
 
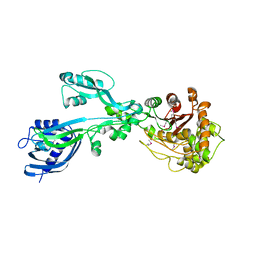 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5EQC
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
5F6R
 
 | | Co-crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) Dehydratase from Yersinia pestis with 5-Benzoylpentanoic Acid | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, 6-oxidanylidene-6-phenyl-hexanoic acid, PHOSPHATE ION | | Authors: | Maltseva, N, Kim, Y, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-06 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.179 Å) | | Cite: | Co-crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) Dehydratase from Yersinia pestis with 5-Benzoylpentanoic Acid
To Be Published
|
|
5F4C
 
 | | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium | | Descriptor: | MALONATE ION, Putative cytoplasmic protein | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Ribonuclease Inhibitor Barstar from Salmonella Typhimurium.
To Be Published
|
|
5E3K
 
 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, Aminotransferase, CARBONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid
To Be Published
|
|
