8GZH
 
 | |
8GZG
 
 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc
Proc.Natl.Acad.Sci.USA, 2023
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7YP9
 
 | |
7YPB
 
 | |
7YPA
 
 | |
6J9E
 
 | |
6J9F
 
 | |
8H3Z
 
 | |
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7EU0
 
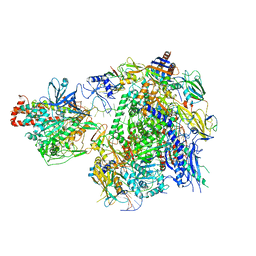 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 backtracked complex | | Descriptor: | DNA (33-MER), DNA (5'-D(*CP*TP*GP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*GP*AP*GP*TP*G)-3'), DNA-directed RNA polymerase IV subunit 1, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
7EU1
 
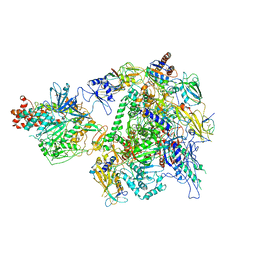 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 holoenzyme | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerases II and IV subunit 5A, DNA-directed RNA polymerases II, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
1ED3
 
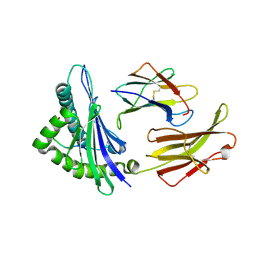 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
