6SXO
 
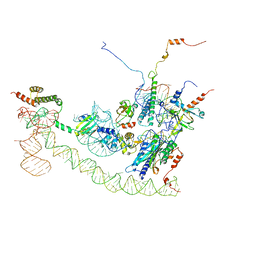 | | Cryo-EM structure of the human Ebp1-ribosome complex | | Descriptor: | 28S ribosomal RNA including ES27L-B (2839-3265), 5.8S ribosomal RNA, 60S ribosomal protein L19, ... | | Authors: | Wild, K, Aleksic, M, Pfeffer, M, Sinning, I. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments.
Nat Commun, 11, 2020
|
|
5L3Q
 
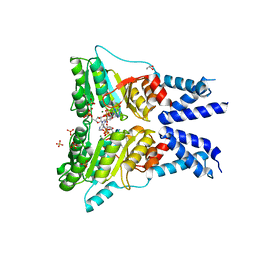 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wild, K, Segnitz, B, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
2VTK
 
 | |
1VTK
 
 | |
3VTK
 
 | |
3KTV
 
 | | Crystal structure of the human SRP19/S-domain SRP RNA complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KTW
 
 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1JID
 
 | | Human SRP19 in complex with helix 6 of Human SRP RNA | | Descriptor: | HELIX 6 OF HUMAN SRP RNA, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN | | Authors: | Wild, K, Sinning, I, Cusack, S. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an early protein-RNA assembly complex of the signal recognition particle.
Science, 294, 2001
|
|
1ZAK
 
 | |
1D4R
 
 | | 29-mer fragment of human srp rna helix 6 | | Descriptor: | 29-MER OF MODIFIED SRP RNA HELIX 6, MAGNESIUM ION | | Authors: | Wild, K, Weichenrieder, O, Leonard, G.A, Cusack, S. | | Deposit date: | 1999-10-05 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2 A structure of helix 6 of the human signal recognition particle RNA
Structure Fold.Des., 7, 1999
|
|
6ZQQ
 
 | | Structure of the Pmt3-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT3 isoform 1 | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
6ZQP
 
 | | Structure of the Pmt2-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
5L3W
 
 | | Structure of the crenarchaeal FtsY GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle receptor FtsY | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3R
 
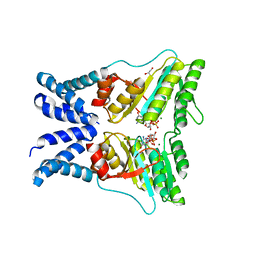 | | Structure of the GTPase heterodimer of chloroplast SRP54 and FtsY from Arabidopsis thaliana | | Descriptor: | Cell division protein FtsY homolog, chloroplastic, GLYCEROL, ... | | Authors: | Bange, G, Kribelbauer, J, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3V
 
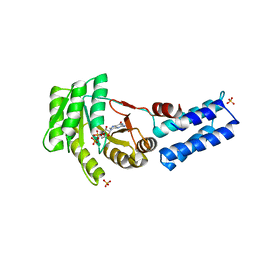 | | Structure of the crenarchaeal SRP54 GTPase bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Signal recognition particle 54 kDa protein | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
5L3S
 
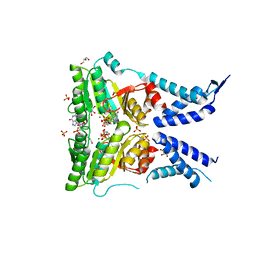 | | Structure of the GTPase heterodimer of crenarchaeal SRP54 and FtsY | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
4JFN
 
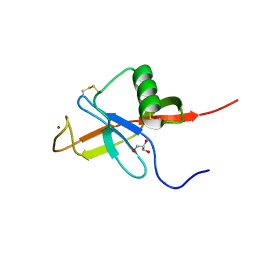 | | Crystal structure of the N-terminal, growth factor-like domain of the amyloid precursor protein bound to copper | | Descriptor: | Amyloid beta A4 protein, COPPER (II) ION, GLYCEROL | | Authors: | Wild, K, Baumkotter, F, Kins, S. | | Deposit date: | 2013-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain
J.Neurosci., 34, 2014
|
|
8QDQ
 
 | |
8QDR
 
 | | Vitis vinifera dimeric 13S-lipoxygenase LOXA in a detergent bound open conformation | | Descriptor: | 1,2-ETHANEDIOL, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, FE (III) ION, ... | | Authors: | Wild, K, Sinning, I. | | Deposit date: | 2023-08-30 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vitis vinifera lipoxygenase LoxA is an allosteric dimer activated by lipidic surfaces.
J.Mol.Biol., 2024
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
7Q72
 
 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q73
 
 | | Structure of Pla1 apo | | Descriptor: | GLYCEROL, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q74
 
 | |
8CQZ
 
 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CR2
 
 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
