5YN8
 
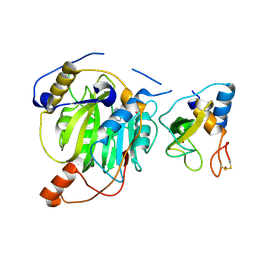 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNJ
 
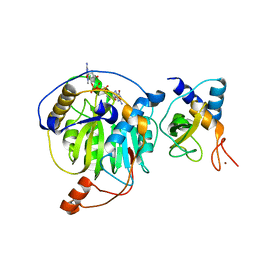 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNO
 
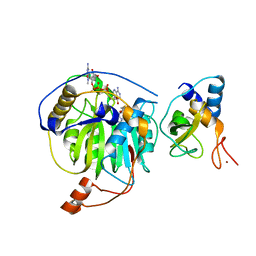 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN6
 
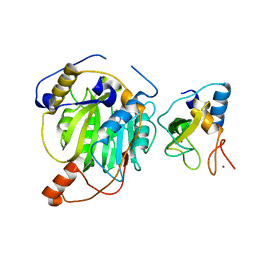 | | Crystal structure of MERS-CoV nsp10/nsp16 complex bound to SAM | | Descriptor: | S-ADENOSYLMETHIONINE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNN
 
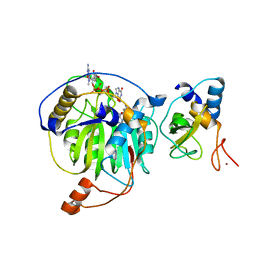 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNI
 
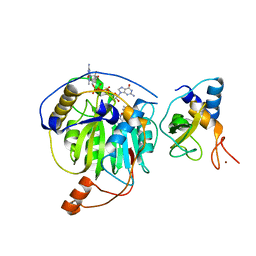 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNQ
 
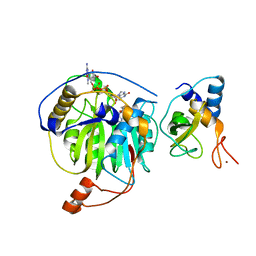 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN5
 
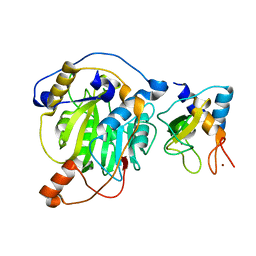 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
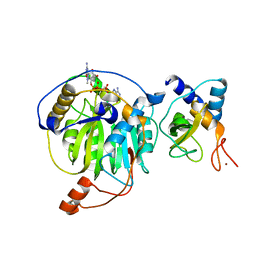 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNP
 
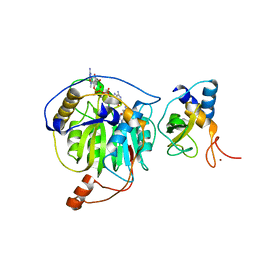 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNB
 
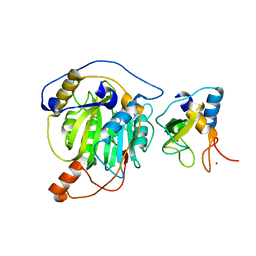 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
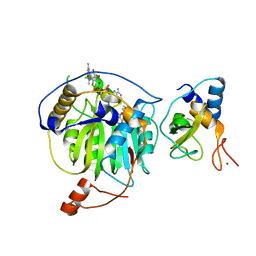 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4EBB
 
 | | Structure of DPP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 2, ZINC ION | | Authors: | Shewchuk, L.M, Hassell, A.H, Sweitzer, S.M, Sweitzer, T.D, McDevitt, P.J, Kennedy-Wilson, K.M, Johanson, K.O. | | Deposit date: | 2012-03-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Human DPP7 Reveal the Molecular Basis of Specific Inhibition and the Architectural Diversity of Proline-Specific Peptidases.
Plos One, 7, 2012
|
|
2YKT
 
 | | Crystal structure of the I-BAR domain of IRSp53 (BAIAP2) in complex with an EHEC derived Tir peptide | | Descriptor: | BRAIN-SPECIFIC ANGIOGENESIS INHIBITOR 1-ASSOCIATED PROTEIN 2, SULFATE ION, TRANSLOCATED INTIMIN RECEPTOR PROTEIN | | Authors: | de Groot, J.C, Schlueter, K, Carius, Y, Quedenau, C, Vingadassalom, D, Faix, J, Weiss, S.M, Reichelt, J, Standfuss-Gabisch, C, Lesser, C.F, Leong, J.M, Heinz, D.W, Buessow, K, Stradal, T.E.B. | | Deposit date: | 2011-05-30 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Complex Formation between Human Irsp53 and the Translocated Intimin Receptor Tir of Enterohemorrhagic E. Coli.
Structure, 19, 2011
|
|
4ZZJ
 
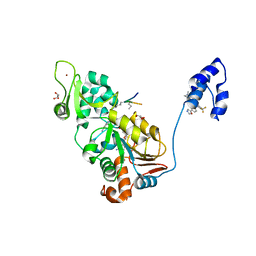 | | SIRT1/Activator/Substrate Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, Ac-p53, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7403 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZH
 
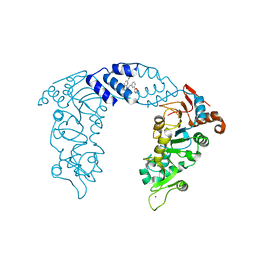 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZI
 
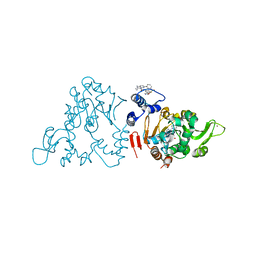 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
