8RW1
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8D
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8G
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8H
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8I
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
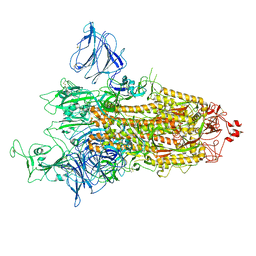 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
