8GLV
 
 | |
6NYS
 
 | |
6M0D
 
 | | Beijerinckia indica beta-fructosyltransferase | | Descriptor: | Levansucrase, MAGNESIUM ION | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6M0E
 
 | | Beijerinckia indica beta-fructosyltransferase complexed with fructose | | Descriptor: | Levansucrase, MAGNESIUM ION, beta-D-fructofuranose, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5B6T
 
 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase complexed with Pb | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
5B6S
 
 | | Catalytic domain of Coprinopsis cinerea GH62 alpha-L-arabinofuranosidase | | Descriptor: | CALCIUM ION, GLYCEROL, Glycosyl hydrolase family 62 protein | | Authors: | Tonozuka, T. | | Deposit date: | 2016-06-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Catalytic Domain of alpha-L-Arabinofuranosidase from Coprinopsis cinerea, CcAbf62A, Provides Insights into Structure-Function Relationships in Glycoside Hydrolase Family 62
Appl. Biochem. Biotechnol., 181, 2017
|
|
6NYR
 
 | |
3VSS
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSR
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
7ZAL
 
 | |
2ZYK
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Tonozuka, T, Sogawa, A, Yamada, M, Matsumoto, N, Yoshida, H, Kamitori, S, Ichikawa, K, Mizuno, M, Nishikawa, A, Sakano, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cyclodextrin recognition by Thermoactinomyces vulgaris cyclo/maltodextrin-binding protein
Febs J., 274, 2007
|
|
5Z0U
 
 | |
5Z0T
 
 | |
1R7R
 
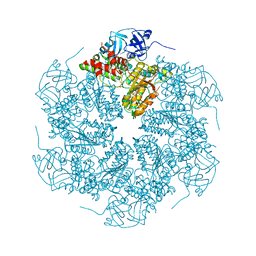 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
2JE0
 
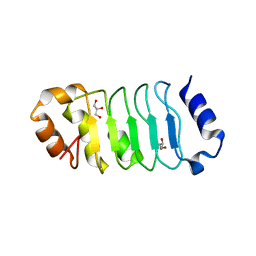 | | Crystal Structure of pp32 | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER A, GLYCEROL | | Authors: | Huyton, T, Wolberger, C. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Tumor Suppressor Protein Pp32 (Anp32A): Structural Insights Into Anp32 Family of Proteins.
Protein Sci., 16, 2007
|
|
2JE1
 
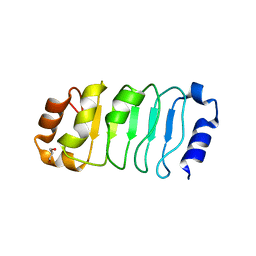 | |
2Q7N
 
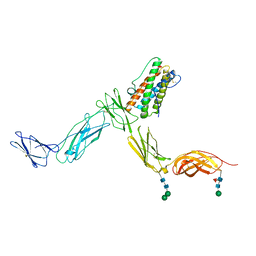 | | Crystal structure of Leukemia inhibitory factor in complex with LIF receptor (domains 1-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huyton, T, Zhang, J.G, Nicola, N.A, Garrett, T.P.J. | | Deposit date: | 2007-06-07 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7KZO
 
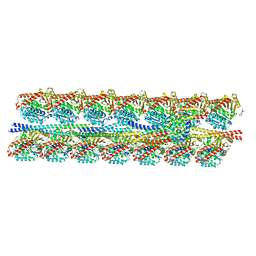 | | Outer dynein arm docking complex bound to doublet microtubules from C. reinhardtii | | Descriptor: | Dynein gamma chain, flagellar outer arm, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
7KZM
 
 | | Outer dynein arm bound to doublet microtubules from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
7KZN
 
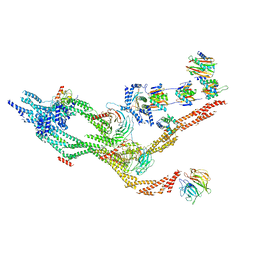 | | Outer dynein arm core subcomplex from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
6QUG
 
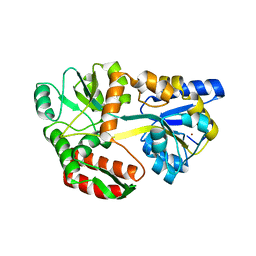 | | GHK tagged MBP-Nup98(1-29) | | Descriptor: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QUI
 
 | | GHK tagged GFP variant at 17Kev | | Descriptor: | COPPER (II) ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8XXA
 
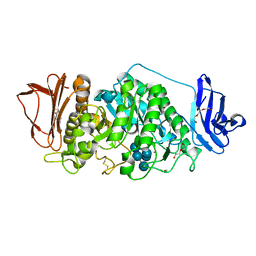 | |
8XX9
 
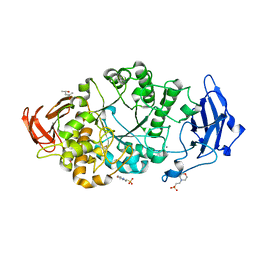 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus.
Proteins, 2024
|
|
8I2Q
 
 | | Beijerinckia indica beta-fructosyltransferase variant H395R/F473Y | | Descriptor: | Beta-fructosyltransferase, GLYCEROL | | Authors: | Tonozuka, T. | | Deposit date: | 2023-01-15 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization and alteration of product specificity of Beijerinckia indica subsp. indica beta-fructosyltransferase.
Biosci.Biotechnol.Biochem., 87, 2023
|
|
