7T0Y
 
 | | The Ribosomal RNA Processing 1B Protein Phosphatase-1 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, FLUORIDE ION, ... | | Authors: | Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The ribosomal RNA processing 1B:protein phosphatase 1 holoenzyme reveals non-canonical PP1 interaction motifs.
Cell Rep, 41, 2022
|
|
8DWL
 
 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8DWK
 
 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8JCR
 
 | | Crystal structure of Procerain from Calotropis gigantea (pH 6.0) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Procerain from Calotropis gigantea
To Be Published
|
|
8JCQ
 
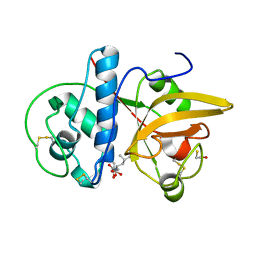 | |
8JCS
 
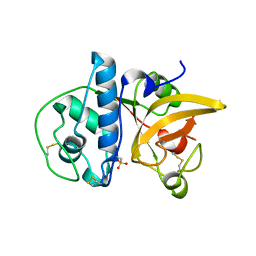 | |
6FGP
 
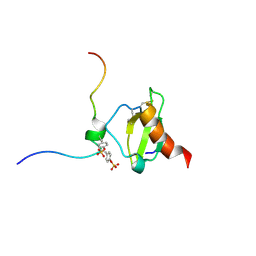 | |
8U5G
 
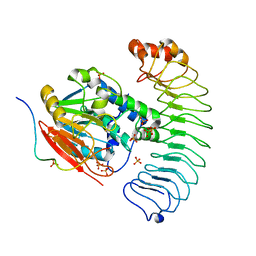 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
