2GNU
 
 | | The crystallization of reaction center from Rhodobacter sphaeroides occurs via a new route | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Wadsten, P, Woehri, A.B, Snijder, A, Katona, G, Gardiner, A.T, Cogdell, R.J, Neutze, R, Engstroem, S. | | Deposit date: | 2006-04-11 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lipidic sponge phase crystallization of membrane proteins
J.Mol.Biol., 364, 2006
|
|
4CTH
 
 | | Neprilysin variant G399V,G714K in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Webster, C.I, Burrell, M, Olsson, L, Fowler, S.B, Digby, S, Sandercock, A, Snijder, A, Tebbe, J, Haupts, U, Grudzinska, J, Jermutus, L, Andersson, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering Neprilysin Activity and Specificity to Create a Novel Therapeutic for Alzheimer'S Disease.
Plos One, 9, 2014
|
|
2BNS
 
 | | Lipidic cubic phase grown reaction centre from Rhodobacter sphaeroides, excited state | | Descriptor: | 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Katona, G, Snijder, A, Gourdon, P, Andreasson, U, Hansson, O, Andreasson, L.E, Neutze, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Regulation of Charge Recombination Reactions in a Photosynthetic Bacterial Reaction Centre
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BNP
 
 | | Lipidic cubic phase grown reaction centre from Rhodobacter sphaeroides, ground state | | Descriptor: | 2-T-BUTYLAMINO-4-ETHYLAMINO-6-METHYLTHIO-S-TRIAZINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Katona, G, Snijder, A, Gourdon, P, Andreasson, U, Hansson, O, Andreasson, L.E, Neutze, R. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Regulation of Charge Recombination Reactions in a Photosynthetic Bacterial Reaction Centre
Nat.Struct.Mol.Biol., 12, 2005
|
|
5LAE
 
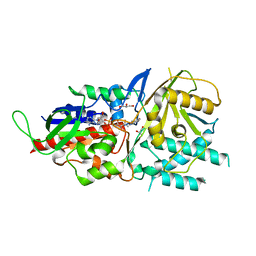 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LFO
 
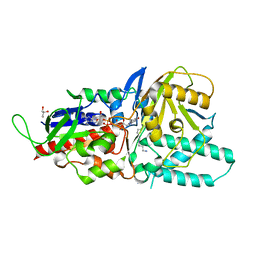 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LGB
 
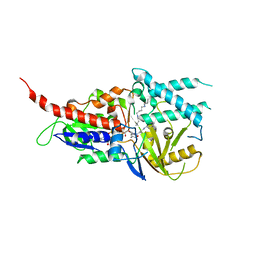 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | Descriptor: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5MBX
 
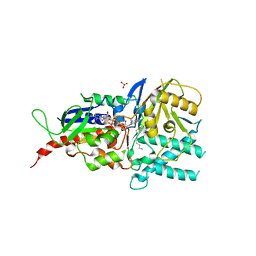 | | Crystal structure of reduced murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
6QZR
 
 | | 14-3-3 sigma in complex with FOXO1 pT24 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Forkhead box protein O1, ... | | Authors: | Ottmann, C, Wolter, M, Lau, R.A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins.
J.Biol.Chem., 294, 2019
|
|
2RDD
 
 | | X-ray crystal structure of AcrB in complex with a novel transmembrane helix. | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Acriflavine resistance protein B, UPF0092 membrane protein yajC | | Authors: | Tornroth-Horsefield, S, Gourdon, P, Horsefield, R, Neutze, R. | | Deposit date: | 2007-09-22 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist.
Structure, 15, 2007
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8ANS
 
 | | Crystal structure of D1228V c-MET bound by compound 1. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, GLYCEROL, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
6YOS
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pT524 pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Glucocorticoid receptor,Glucocorticoid receptor | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
6YMO
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pS617 peptide | | Descriptor: | 14-3-3 protein zeta/delta, 2-HYDROXYBENZOIC ACID, Glucocorticoid receptor, ... | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
6YO8
 
 | | Binary complex of 14-3-3 zeta with Glucocorticoid Receptor (GR) pT524 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Glucocorticoid receptor | | Authors: | Munier, C.C, Edman, K, Perry, M.W.D, Ottmann, C. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators.
J.Biol.Chem., 296, 2021
|
|
6QZS
 
 | | 14-3-3 sigma in complex with FOXO1 pS256 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FOXO1 pS256 site, ... | | Authors: | Ottmann, C, Wolter, M, Lau, R.A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins.
J.Biol.Chem., 294, 2019
|
|
5JNQ
 
 | | MraY tunicamycin complex | | Descriptor: | PALMITIC ACID, Phospho-N-acetylmuramoyl-pentapeptide-transferase, Tunicamycin, ... | | Authors: | Johansson, P. | | Deposit date: | 2016-04-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MraY-antibiotic complex reveals details of tunicamycin mode of action.
Nat. Chem. Biol., 13, 2017
|
|
6SD9
 
 | | Crystal structure of wild-type cMET bound by foretinib | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDC
 
 | | Crystal structure of D1228V cMET bound by foretinib | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDD
 
 | | Crystal structure of D1228V cMET bound by BMS-777607 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-{4-[(2-amino-3-chloropyridin-4-yl)oxy]-3-fluorophenyl}-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Collie, G.W, Phillips, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Molecular Insight into Resistance Mechanisms of First Generation cMET Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6SDE
 
 | |
7B42
 
 | | Crystal structure of c-MET bound by compound 8 | | Descriptor: | 3-[(3-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
