1MT0
 
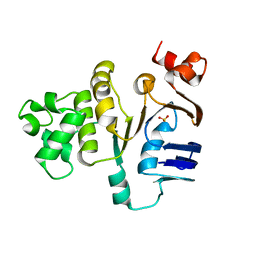 | | ATP-binding domain of hemolysin B from Escherichia coli | | Descriptor: | Hemolysin secretion ATP-binding protein, SULFATE ION | | Authors: | Schmitt, L, Benabdelhak, H, Blight, M.A, Holland, I.B, Stubbs, M.T. | | Deposit date: | 2002-09-20 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the nucleotide binding domain of the
ABC-transporter hemolysin B: identification of a variable region
within ABC helical domains
J.Mol.Biol., 330, 2003
|
|
4MEE
 
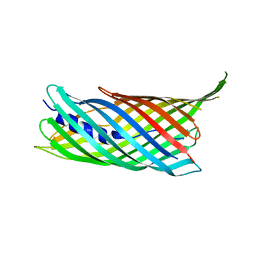 | | Crystal structure of the transport unit of the autotransporter AIDA-I from Escherichia coli | | Descriptor: | Diffuse adherence adhesin | | Authors: | Gawarzewski, I, Tschapek, B, Hoeppner, A, Smits, S.H, Jose, J, Schmitt, L. | | Deposit date: | 2013-08-26 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transport unit of the autotransporter adhesin involved in diffuse adherence from Escherichia coli.
J.Struct.Biol., 187, 2014
|
|
8BYK
 
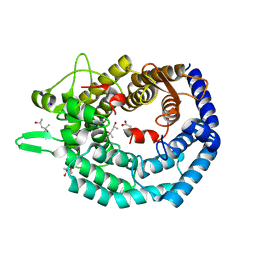 | | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Knospe, C.V, Kamel, M, Spitz, O, Hoeppner, A, Galle, S, Reiners, J, Kedrov, A, Smits, S.H, Schmitt, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases.
Front Microbiol, 13, 2022
|
|
6EYL
 
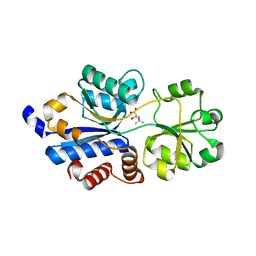 | | Crystal structure of OpuBC in complex with carnitine | | Descriptor: | CARNITINE, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Peherstorfer, S, Teichmann, L, Smits, S.H, Sschmitt, L, Bremer, E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reprogramming the substrate specificity of an ABC import system by a single amino acid substitution in its cognate ligand binding protein
To Be Published
|
|
3QHQ
 
 | | Structure of CRISPR-associated protein Csn2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Sag0897 family CRISPR-associated protein | | Authors: | Ellinger, P, Arslan, Z, Wurm, R, Tschapek, B, Pfeffer, K, Wagner, R, Schmitt, L, Pul, U, Smits, S.H. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of CRISPR-associated protein Csn2
To be Published
|
|
3R6U
 
 | | Crystal structure of choline binding protein OpuBC from Bacillus subtilis | | Descriptor: | CHOLINE ION, Choline-binding protein | | Authors: | Pittelkow, M, Tschapek, B, Smits, S.H.J, Schmitt, L, Bremer, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of the Substrate-Binding Protein OpuBC from Bacillus subtilis in Complex with Choline.
J.Mol.Biol., 411, 2011
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7PZE
 
 | | MademoiseLLE domain 2 of Rrm4 from Ustilago maydis | | Descriptor: | Chromosome 8, whole genome shotgun sequence | | Authors: | Devans, S, Schott-Verdugo, s, Muentjes, K, Olgeiser, L, Reiners, J, Schmitt, L, Hoeppner, A, Smits, S.H, Gohlke, H, Feldbruegge, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A MademoiseLLE domain binding platform links the key RNA transporter to endosomes.
Plos Genet., 18, 2022
|
|
8P0S
 
 | | Crystal structure HR1 domain of Rho-associated coiled-coil protein kinases (ROCK-HR1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Rho-associated protein kinase 1 | | Authors: | Dubey, B.N, Dvorsky, R, Gremer, L, Vetter, I.R, Schmitt, L, Groth, G, Ahmadian, M.R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the p160 Rho-associated coiled-coil-containing protein kinase
To Be Published
|
|
1XEF
 
 | | Crystal structure of the ATP/Mg2+ bound composite dimer of HlyB-NBD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB, MAGNESIUM ION | | Authors: | Zaitseva, J, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2004-09-10 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB
Embo J., 24, 2005
|
|
2REJ
 
 | | ABC-transporter choline binding protein in unliganded semi-closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-26 | | Release date: | 2008-09-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2RF1
 
 | | Crystal structure of ChoX in an unliganded closed conformation | | Descriptor: | PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
2RIN
 
 | | ABC-transporter choline binding protein in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
3MAM
 
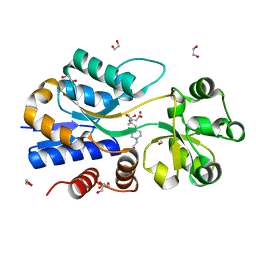 | | A molecular switch changes the low to the high affinity state in the substrate binding protein AfProX | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Osmoprotection protein (ProX), ... | | Authors: | Tschapek, B, Pittelkow, M, Bremer, E, Schmitt, L, Smits, S.H. | | Deposit date: | 2010-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arg149 Is Involved in Switching the Low Affinity, Open State of the Binding Protein AfProX into Its High Affinity, Closed State.
J.Mol.Biol., 411, 2011
|
|
2FGJ
 
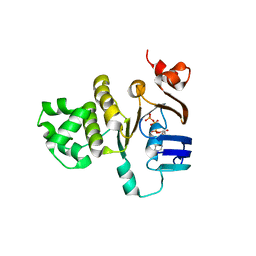 | | Crystal structure of the ABC-cassette H662A mutant of HlyB with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
2FFB
 
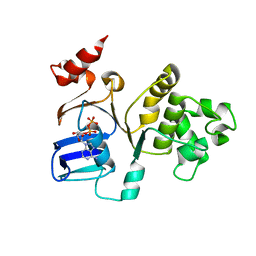 | | The crystal structure of the HlyB-NBD E631Q mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-19 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
2FFA
 
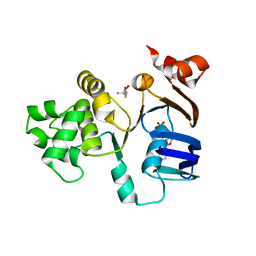 | | Crystal structure of ABC-ATPase H662A of the ABC-transporter HlyB in complex with ADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-19 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
2FGK
 
 | | Crystal structure of the ABC-cassette E631Q mutant of HlyB with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
2FF7
 
 | | The ABC-ATPase of the ABC-transporter HlyB in the ADP bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-hemolysin translocation ATP-binding protein hlyB | | Authors: | Zaitseva, J, Oswald, C, Jumpertz, T, Jenewein, S, Holland, I.B, Schmitt, L. | | Deposit date: | 2005-12-19 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer.
Embo J., 25, 2006
|
|
2Q88
 
 | | Crystal structure of EhuB in complex with ectoine | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
2Q89
 
 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
2B4L
 
 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
