2VN3
 
 | | Nitrite Reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
7CMG
 
 | | The Structure of the periplasmic domain of PorM | | Descriptor: | Por secretion system protein porM/gldM | | Authors: | Sato, K, Okada, K, Nakayam, K, Imada, K. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | PorM, a core component of bacterial type IX secretion system, forms a dimer with a unique kinked-rod shape.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
2VMJ
 
 | | Type 1 Copper-Binding Loop of Nitrite Reductase mutant: 130- CAPEGMVPWHVVSGM-144 to 130-CTPHPFM-136 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
1K37
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
1K36
 
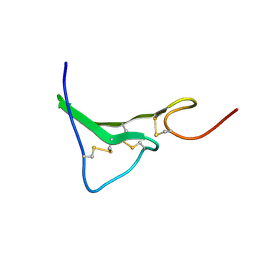 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
2Z0D
 
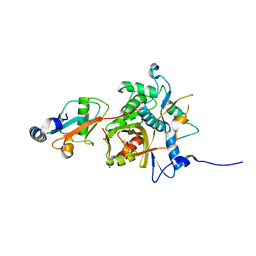 | |
2Z0E
 
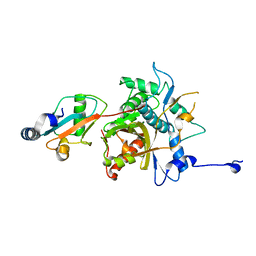 | |
2ZZP
 
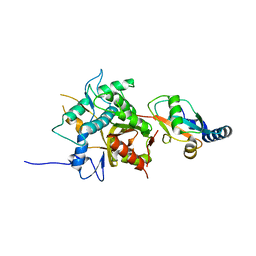 | |
7E8D
 
 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
5Y1A
 
 | | HBP35 of Porphyromonas gingivalis | | Descriptor: | 35 kDa hemin binding protein | | Authors: | Kakuda, S, Suzuki, M, Sato, K. | | Deposit date: | 2017-07-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Immunoglobulin-like domains of the cargo proteins are essential for protein stability during secretion by the type IX secretion system.
Mol. Microbiol., 110, 2018
|
|
2A6W
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), metal-free form | | Descriptor: | Emp46p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A71
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), orthorhombic crystal form | | Descriptor: | Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Y
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), tetragonal crystal form | | Descriptor: | Emp47p (form1), SULFATE ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6V
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), potassium-bound form | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6Z
 
 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 1 | | Descriptor: | Emp47p (form2) | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
2A6X
 
 | | Crystal structure of Emp46p carbohydrate recognition domain (CRD), Y131F mutant | | Descriptor: | 1,2-ETHANEDIOL, Emp46p, POTASSIUM ION | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Kato, R, Nakano, A, Wakatsuki, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
8JUF
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUD
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUG
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
1D6Q
 
 | | HUMAN LYSOZYME E102 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL GLYCOSIDE OF N-ACETYLLACTOSAMINE | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
1D6P
 
 | | HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LYSOZYME | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
